 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN538944.1_FGP027 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 512aa MW: 53504.1 Da PI: 5.2411 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 15.4 | 5.4e-05 | 66 | 83 | 2 | 19 |
EETTTTEEESSHHHHHHH CS
zf-C2H2 2 kCpdCgksFsrksnLkrH 19
+C++Cgk F++++++ H
KN538944.1_FGP027 66 VCKNCGKEFTSWEHFLEH 83
7**************999 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 140 | 65 | 85 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.141 | 65 | 92 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 4.06E-8 | 223 | 246 | No hit | No description |
| SMART | SM00355 | 0.13 | 224 | 246 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF13912 | 2.8E-9 | 224 | 248 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 10.242 | 224 | 251 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 226 | 246 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 4.06E-8 | 332 | 359 | No hit | No description |
| SMART | SM00355 | 3.7 | 337 | 359 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.182 | 337 | 359 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF13912 | 4.2E-11 | 337 | 360 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 339 | 359 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 512 aa Download sequence Send to blast |
MRAHGVGDGD GLGADDDDSL GDEAVRRARG GADDPWNAGG PSSSGAATHV YALRTNPNRV 60 TRSRQVCKNC GKEFTSWEHF LEHGKCSSGE DDDDEDDADR SLQPWSPSPD ADGEEDPAPA 120 AGWLKGKRSR RCKGTGVDLS PTPSACAAGE EEDLANCLVM LSSSKVDQAG VTEAEQPSSS 180 SASKEHKRLI TFMEPTTYVL DTVMALPPPA PAPQYVSTVP RGMFECKACK KVFSSHQALG 240 GHRASHKKVK GCFAAKLESN AAEVAEPSHH AEVADRSEDN PGKATSDARR NVHASMDGDA 300 NAGTSDAAAE LSMAIVPIEP PVAALAAAPL KKKDKMHECS VCHRLFTSGQ ALGGHKRCHW 360 LTSSSADHTA SVPPLADDLV PLSFRPMLDA PEPALDLSIA ANPPLLASAA TVRPKVGGSS 420 FHLDAPPPVY IPSSPAIPSQ RNKATATTGS QNANDAVGLS TAAAEDEADS TTVKRARLSD 480 LKDVSMAGET TPWLQVGIGS SSRGGGDDND KE |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00621 | PBM | Transfer from AT2G17180 | Download |
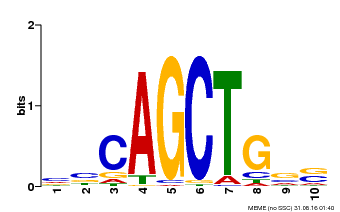
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN538944.1_FGP027 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012612 | 0.0 | CP012612.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 4 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015634601.1 | 0.0 | uncharacterized protein LOC4336604 | ||||
| TrEMBL | I1PNN8 | 0.0 | I1PNN8_ORYGL; Uncharacterized protein | ||||
| STRING | ORGLA04G0180700.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3524 | 32 | 62 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G17180.1 | 3e-20 | C2H2-like zinc finger protein | ||||



