 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN538954.1_FGP008 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 398aa MW: 42759.1 Da PI: 8.4199 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.1 | 2.8e-12 | 159 | 198 | 11 | 55 |
HHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 11 rRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
rRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
KN538954.1_FGP008 159 RRRDRINEKMRALQELIPNC-----NKIDKASMLDEAIEYLKTLQ 198
8******************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 16.107 | 148 | 197 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.88E-16 | 149 | 207 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.1E-16 | 151 | 206 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.29E-13 | 152 | 202 | No hit | No description |
| SMART | SM00353 | 1.0E-13 | 155 | 203 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.0E-9 | 159 | 198 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 398 aa Download sequence Send to blast |
MAKLELSCGK KKKEKSLVMR TATNCSRSDG NDFAELLWEN GQAVVHGRKK HPQPAFPPFG 60 FFGGTGGGGG GSSSRAQERQ PGGIDAFAKA RKAPEATVAT SSVCSGNGAG SDELWRQQKR 120 KCQAQAECSA SQDDDLDDEP GVLRKSGTRS TKRSRTAERR RDRINEKMRA LQELIPNCNK 180 IDKASMLDEA IEYLKTLQLQ VQMMSMGTGL CIPPMLLPTA MQHLQIPPMA HFPHLGMGLG 240 YGMGVFDMSN TGALQMPPMP GAHFPCPMIP GASPQGLGIP GTSTMPMFGV PGQTIPSSAY 300 SVPPFASLAG LPVRPSGVPQ VSGAMANMVQ DQQQGIANQQ QQCLNKEAIQ GANPGDSQMQ 360 IIMQNVPVLK RSMAEEFHMV EDATRLQAVI LDFQCSFL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction. {ECO:0000269|PubMed:17485859}. | |||||
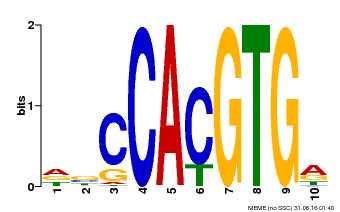
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN538954.1_FGP008 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light in dark-grown etiolated seedlings. {ECO:0000269|PubMed:17485859}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK102252 | 0.0 | AK102252.1 Oryza sativa Japonica Group cDNA clone:J033088H11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015619034.1 | 1e-175 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15-like isoform X2 | ||||
| Swissprot | Q0JNI9 | 1e-176 | PIL15_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15 | ||||
| TrEMBL | A0A0E0MUV1 | 0.0 | A0A0E0MUV1_ORYRU; Uncharacterized protein | ||||
| STRING | ORUFI01G12840.1 | 0.0 | (Oryza rufipogon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-28 | phytochrome interacting factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



