 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN538979.1_FGP004 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 571aa MW: 62956.9 Da PI: 10.5834 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 396.8 | 2.9e-121 | 268 | 571 | 15 | 301 |
GAGA_bind 15 epaaslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllalllvensla...salpvgvqvls 103
+ +++lk++++++l++++++rd++irer++al+ekkaa+aerdmaf+qrd+a+aern+a+verdn+l+al+l++++ + +++ +++ls
KN538979.1_FGP004 268 QTQRHLKDHQSMNLLALMNDRDNAIRERDHALAEKKAAIAERDMAFTQRDAAMAERNAAVVERDNALAALELARTNGLnmnNGNGFPQGSLS 359
44589*****************************************************************99888765566889999***** PP
GAGA_bind 104 gtksidslqqlse.....pqledsave.lreeeklealpieeaaeeakekkkkkkrqrakkpkekkak..kkkkksekskkkvkkesader. 186
g+k+i++++qls+ ql+ds+++ +re++++ea+pi++a+ +a ++k++kk++++++p ++ + +k+kk++ ++k+v ++++ ++
KN538979.1_FGP004 360 GSKNIHHHDQLSHaqsspLQLADSPYDhTREMHISEAYPISTAPGSAGKAKRPKKNSSQASPSKRPSGvlRKTKKPSGDWKNVGMSGCGDDs 451
**********888899999********9***********************************9999988888889**********998555 PP
GAGA_bind 187 .....skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagrKmSqgafkklLekLa 273
+k+e+k+++l+ln+v++Dest+P+P+CsCtG+lrqCYkWGnGGWqS+CCt++iS+yPLPv++++r+aR++grKmS+gaf+klL++La
KN538979.1_FGP004 452 ahasvMKNEWKDQNLGLNQVAFDESTMPAPACSCTGKLRQCYKWGNGGWQSSCCTMNISMYPLPVMPNKRHARMGGRKMSGGAFTKLLSRLA 543
89999*************************************************************************************** PP
GAGA_bind 274 aeGydlsnpvDLkdhWAkHGtnkfvtir 301
aeG+dls+pvDLkdhWAkHGtn+++tir
KN538979.1_FGP004 544 AEGHDLSTPVDLKDHWAKHGTNRYITIR 571
***************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00125 | 1.3E-32 | 1 | 102 | IPR007125 | Histone H2A/H2B/H3 |
| Gene3D | G3DSA:1.10.20.10 | 8.9E-58 | 2 | 101 | IPR009072 | Histone-fold |
| SuperFamily | SSF47113 | 7.18E-37 | 2 | 102 | IPR009072 | Histone-fold |
| PRINTS | PR00622 | 1.7E-55 | 3 | 17 | IPR000164 | Histone H3/CENP-A |
| PROSITE pattern | PS00322 | 0 | 15 | 21 | IPR000164 | Histone H3/CENP-A |
| PRINTS | PR00622 | 1.7E-55 | 17 | 31 | IPR000164 | Histone H3/CENP-A |
| SMART | SM00428 | 6.8E-29 | 34 | 122 | IPR000164 | Histone H3/CENP-A |
| PRINTS | PR00622 | 1.7E-55 | 34 | 55 | IPR000164 | Histone H3/CENP-A |
| PRINTS | PR00622 | 1.7E-55 | 58 | 75 | IPR000164 | Histone H3/CENP-A |
| PROSITE pattern | PS00959 | 0 | 67 | 75 | IPR000164 | Histone H3/CENP-A |
| PRINTS | PR00622 | 1.7E-55 | 80 | 98 | IPR000164 | Histone H3/CENP-A |
| Gene3D | G3DSA:1.10.10.10 | 2.2E-17 | 148 | 228 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 2.56E-15 | 149 | 228 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| CDD | cd00073 | 1.77E-19 | 149 | 222 | No hit | No description |
| SMART | SM00526 | 4.6E-25 | 149 | 218 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 24.096 | 151 | 223 | IPR005818 | Linker histone H1/H5, domain H15 |
| Pfam | PF00538 | 1.0E-11 | 152 | 221 | IPR005818 | Linker histone H1/H5, domain H15 |
| SMART | SM01226 | 3.4E-165 | 253 | 571 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 2.1E-96 | 255 | 571 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 571 aa Download sequence Send to blast |
MARTKQTARK STGGKAPRKQ LATKAARKSA PTTGGVKKPH RYRPGTVALR EIRKYQKSTE 60 LLIRKLPFQR LVREIAQDFK TDLRFQSHAV LALQEAAEAY LEVKEAVEAP KPEEAPKAAE 120 EVEEKKAEGE KEKAKKERKP RARKPRSAGP HHPPYFEMIK EAIMALDGNG KAGSSPYAIA 180 KYMGEQHMGV LPANYRKVLA VQLRNFAAKG RLVKVKASFK LSAAEEKKAT AASVGFGVPA 240 AIKPTEEKHA RWDDERVSAS ASMWMMPQTQ RHLKDHQSMN LLALMNDRDN AIRERDHALA 300 EKKAAIAERD MAFTQRDAAM AERNAAVVER DNALAALELA RTNGLNMNNG NGFPQGSLSG 360 SKNIHHHDQL SHAQSSPLQL ADSPYDHTRE MHISEAYPIS TAPGSAGKAK RPKKNSSQAS 420 PSKRPSGVLR KTKKPSGDWK NVGMSGCGDD SAHASVMKNE WKDQNLGLNQ VAFDESTMPA 480 PACSCTGKLR QCYKWGNGGW QSSCCTMNIS MYPLPVMPNK RHARMGGRKM SGGAFTKLLS 540 RLAAEGHDLS TPVDLKDHWA KHGTNRYITI R |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3lel_A | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 3lel_E | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 3lel_K | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 3lel_O | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 4zux_A | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 4zux_E | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 4zux_K | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 4zux_O | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 5b40_A | 3e-59 | 1 | 101 | 4 | 104 | Histone H3.2 |
| 5b40_E | 3e-59 | 1 | 101 | 4 | 104 | Histone H3.2 |
| 5e5a_A | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 5e5a_E | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 5o9g_A | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 5o9g_E | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 6i84_M | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 6i84_S | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 6j99_A | 3e-59 | 1 | 101 | 1 | 101 | Histone H3 |
| 6j99_E | 3e-59 | 1 | 101 | 1 | 101 | Histone H3 |
| 6ne3_A | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| 6ne3_E | 3e-59 | 1 | 101 | 1 | 101 | Histone H3.2 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
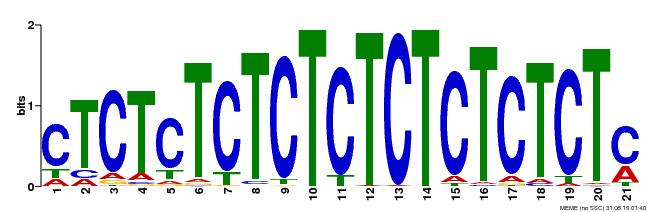
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN538979.1_FGP004 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012614 | 0.0 | CP012614.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 6 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015641153.1 | 0.0 | barley B recombinant-like protein D | ||||
| Refseq | XP_015641154.1 | 0.0 | barley B recombinant-like protein D | ||||
| Swissprot | Q5VSA8 | 0.0 | BBRD_ORYSJ; Barley B recombinant-like protein D | ||||
| TrEMBL | A0A0D3GCC3 | 0.0 | A0A0D3GCC3_9ORYZ; Uncharacterized protein | ||||
| TrEMBL | A0A0E0A4I0 | 0.0 | A0A0E0A4I0_9ORYZ; Uncharacterized protein | ||||
| TrEMBL | A0A0E0HKD4 | 0.0 | A0A0E0HKD4_ORYNI; Uncharacterized protein | ||||
| TrEMBL | I1PZ79 | 0.0 | I1PZ79_ORYGL; Uncharacterized protein | ||||
| STRING | OGLUM06G01860.1 | 0.0 | (Oryza glumipatula) | ||||
| STRING | ONIVA06G02210.1 | 0.0 | (Oryza nivara) | ||||
| STRING | ORGLA06G0019100.1 | 0.0 | (Oryza glaberrima) | ||||
| STRING | OBART06G01840.1 | 0.0 | (Oryza barthii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6226 | 37 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 9e-94 | basic pentacysteine 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



