 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN539031.1_FGP022 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 112aa MW: 12588.1 Da PI: 8.5278 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 62.3 | 1e-19 | 11 | 56 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEde l ++v+++G ++W++I r ++ gR++k+c++rw +
KN539031.1_FGP022 11 KGPWSPEEDEALRRLVERHGARNWTAIGRGIP-GRSGKSCRLRWCNQ 56
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 41.4 | 3.2e-13 | 63 | 98 | 1 | 38 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlk 38
r ++T+eEd +++a+++lG++ W++Iar ++ gRt++
KN539031.1_FGP022 63 RRPFTAEEDAAILRAHARLGNR-WAAIARLLP-GRTDN 98
679*******************.*********.****9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 25.481 | 6 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.64E-29 | 8 | 99 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.6E-17 | 10 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.2E-19 | 11 | 56 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.7E-26 | 12 | 64 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.48E-15 | 13 | 55 | No hit | No description |
| SMART | SM00717 | 3.4E-5 | 62 | 110 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.0E-11 | 63 | 99 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 9.793 | 63 | 103 | IPR017930 | Myb domain |
| CDD | cd00167 | 3.87E-8 | 65 | 99 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-17 | 65 | 99 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:2000022 | Biological Process | regulation of jasmonic acid mediated signaling pathway | ||||
| GO:2000031 | Biological Process | regulation of salicylic acid mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 112 aa Download sequence Send to blast |
MGVEAECDRI KGPWSPEEDE ALRRLVERHG ARNWTAIGRG IPGRSGKSCR LRWCNQLSPQ 60 VERRPFTAEE DAAILRAHAR LGNRWAAIAR LLPGRTDNAS RGEVEERGDG EM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-33 | 8 | 103 | 4 | 99 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor (PubMed:23067202, PubMed:23603962). Represses the expression of protein phosphatases 2C in response to abscisic acid (ABA). Confers resistance to abiotic stresses dependent of ABA (PubMed:18162593, PubMed:9678577). In response to auxin, activates the transcription of the auxin-responsive gene IAA19. The IAA19 transcription activation by MYB44 is enhanced by direct interaction between MYB44 and PYL8 (PubMed:24894996). Transcriptional activator of WRKY70 by direct binding to its promoter region, especially at 5'-TAACNG-3' and 5'-CNGTTA-3' symmetric motifs (PubMed:23067202, PubMed:23603962). Activates salicylic acid (SA)- mediated defenses and subsequent resistance to biotrophic pathogen P.syringae pv. tomato DC3000, but represses jasmonic acid (JA)-mediated defenses responses against the necrotrophic pathogen A.brassicicola (PubMed:23067202). {ECO:0000269|PubMed:18162593, ECO:0000269|PubMed:23067202, ECO:0000269|PubMed:23603962, ECO:0000269|PubMed:24894996, ECO:0000269|PubMed:9678577}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00653 | PBM | Transfer from LOC_Os02g09480 | Download |
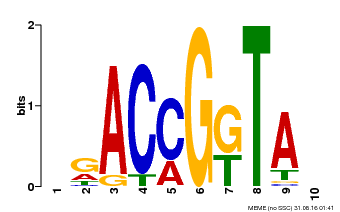
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN539031.1_FGP022 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By drought, cold, high salinity, cadmium (CdCl(2)), salicylic acid (SA), jasmonate (JA), ethylene, gibberellic acid (GA), and ABA (PubMed:16463103, PubMed:18162593, PubMed:23067202). The induction by JA is COI1-dependent (PubMed:23067202). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:18162593, ECO:0000269|PubMed:23067202}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP003990 | 1e-160 | AP003990.3 Oryza sativa Japonica Group genomic DNA, chromosome 2, BAC clone:OJ1073_F05. | |||
| GenBank | AP004046 | 1e-160 | AP004046.2 Oryza sativa Japonica Group genomic DNA, chromosome 2, BAC clone:OJ1145_F01. | |||
| GenBank | AP014958 | 1e-160 | AP014958.1 Oryza sativa Japonica Group DNA, chromosome 2, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015623498.1 | 2e-64 | transcription factor MYB44 | ||||
| Swissprot | Q9FDW1 | 6e-49 | MYB44_ARATH; Transcription factor MYB44 | ||||
| TrEMBL | A0A0E0JWK1 | 7e-64 | A0A0E0JWK1_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC02G05820.1 | 1e-64 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP789 | 38 | 154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G67300.1 | 2e-51 | myb domain protein r1 | ||||



