 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN539406.1_FGP004 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 1441aa MW: 157146 Da PI: 6.5231 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 37.2 | 7.4e-12 | 240 | 296 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng.krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d + +g rk + g ++t+++Aa+a++ a++k++g
KN539406.1_FGP004 240 SQYRGVTRHRWTGRYEAHLWDnSCRkDGqTRKGRQ-GGYDTEDKAARAYDLAALKYWG 296
78*******************54445554335555.77***99*************97 PP
| |||||||
| 2 | AP2 | 45.5 | 1.9e-14 | 339 | 390 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV+++++ grW A+I + +k +lg+f t eeAa+a++ a+ k++g
KN539406.1_FGP004 339 SIYRGVTRHHQHGRWQARIGRVAG---NKDLYLGTFSTQEEAAEAYDIAAIKFRG 390
57****************988532...5************************997 PP
| |||||||
| 3 | HLH | 34.1 | 4.8e-11 | 1299 | 1347 | 3 | 55 |
HHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 3 rahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++iN+++ L++l+P + +k Ka++L + ++Y++sLq
KN539406.1_FGP004 1299 NSHSLAERFRREKINERMKLLQDLVPGC----NKITGKAMMLDEIINYVQSLQ 1347
58**************************....999*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 3.3E-9 | 240 | 296 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.58E-18 | 240 | 304 | No hit | No description |
| SuperFamily | SSF54171 | 7.85E-14 | 240 | 306 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 17.662 | 241 | 304 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.5E-21 | 241 | 310 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.7E-11 | 241 | 305 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.2E-5 | 242 | 253 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.49E-24 | 339 | 400 | No hit | No description |
| Pfam | PF00847 | 2.4E-9 | 339 | 390 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.99E-17 | 339 | 399 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.3E-33 | 340 | 404 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.045 | 340 | 398 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.9E-17 | 340 | 398 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.2E-5 | 380 | 400 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.10.20.90 | 4.1E-5 | 555 | 630 | No hit | No description |
| PROSITE profile | PS50181 | 11.405 | 868 | 914 | IPR001810 | F-box domain |
| SuperFamily | SSF81383 | 2.35E-13 | 868 | 937 | IPR001810 | F-box domain |
| Gene3D | G3DSA:1.20.1280.50 | 1.1E-10 | 871 | 937 | No hit | No description |
| Pfam | PF12937 | 8.9E-6 | 873 | 915 | IPR001810 | F-box domain |
| SMART | SM00256 | 1.5E-5 | 874 | 914 | IPR001810 | F-box domain |
| SuperFamily | SSF47459 | 9.16E-17 | 1294 | 1362 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.30E-11 | 1294 | 1351 | No hit | No description |
| PROSITE profile | PS50888 | 15.796 | 1296 | 1346 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.5E-17 | 1296 | 1363 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.6E-8 | 1299 | 1347 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.3E-10 | 1302 | 1352 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1441 aa Download sequence Send to blast |
MSSPPPDSAT TCNFLFSPPA AQMVAPSPGY YYVGGAYGDG TSTAGVYYSH LPVMPIKSDG 60 SLCIMEGMMP SSSPKLEDFL GCGNGSGHDP ATYYSQGQEA EDAGRAAYQH HQLVPYNYQP 120 LTEAEMLQEA AAAPMEDAMA AAKNFLVTSY GACYGNQEMP QPLSLSMSPG SQSSSCVSAA 180 PQQHQQMAVV AAAAAAGDGQ GSNSNDGGEQ RVGKKRGTGK GGQKQPVHRK SIDTFGQRTS 240 QYRGVTRHRW TGRYEAHLWD NSCRKDGQTR KGRQGGYDTE DKAARAYDLA ALKYWGLSTH 300 INFPLENYRD EIEEMERMTR QEYVAHLRRR SSGFSRGASI YRGVTRHHQH GRWQARIGRV 360 AGNKDLYLGT FSTQEEAAEA YDIAAIKFRG LNAVTNFDIT RYDVDKIMES SSLLPGEAAR 420 KVKAIEAAPE HVPVGRELGA TEEASAATVM GTDWRMVLHG SQQQQAAACT EATADLQKGF 480 MGDAHSALHG IVGFDVESAA ADEIDWGRGE FTAGERCHKA SYIPAPIIPK GFSCRFPRQC 540 FHVPACSFMM AWPVKLPDTA TLSDVKAFLA TKLSAAQPVP AESVRLTLNR SEELLTPDPS 600 ATLPALGLAS GDLLYFTLSP LPSPSPPPQP QPQAQPLPRN PNPDVPSIAG AAGPTKSPVE 660 SGSSSSMPQA LCTNPGLPVT SDPHPPPPDV VMAEAFAAIK SKSSFLVGAM KREMENAGGA 720 DGTVICRLVV ALHAALLDAG FLYANPVGSC LQLPQNWASG SFVPVSMKYT LPELVEALPA 780 VEEGMVAVLN YSLMGNFMMV YGHVPGATSG VRRLCLELPE LAPLLYLDSD EVSTAEEREI 840 HKLWRVLKDD MCLPLMISLC QLNNLSLPPC LMALPGDVKA KVLEFVPGVD LARVQCTCKE 900 LRDLAADDNL WKKKCEMEFN TQGESSRVGR NWKESFYETM STVRFCFDQA FSARTFGCAH 960 EHDSETCVLM IRYMRCMMCK CIYSDQRKDI ILADKYTRGN YMQKPVTQPG RWFILLVFHS 1020 LLCQYIVIGL SLLWYHLVDL VQDAPAAGIH FDCIIPLPIN PYQLLPSAGA CCSTTQASAS 1080 AKDGGNWIAV ASVPWFGLHF TVPTAIDLNS ENFILIHGCD PAGAWLLAVP TILFQDWFPP 1140 LPLRLSIALG LLNFSLWTIF ICIPLPAVLL QAAKGIVSRS ELTSLLRLTL PWTSTSLSFF 1200 LCIMQVCKRG FSFTILPVFP CSNSPDTQLS RNMSIDKCLK GSKRKGSGEG SSSLHSQEET 1260 GEMPQRELSM EHAGEKAGDA DASREEYVHV RAKRGQATNS HSLAERFRRE KINERMKLLQ 1320 DLVPGCNKIT GKAMMLDEII NYVQSLQRQV EFLSMKLSTI SPELNSDLDL QDILCSQDAR 1380 SAFLGCSPQL SNAHPNLYRA AQQCLSPPGL YGSVCVPNPA DVHLARAAHL ASFPQVYIEL 1440 Q |
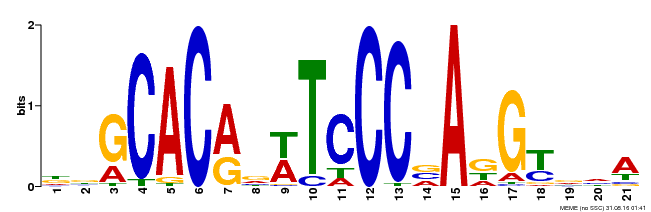
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN539406.1_FGP004 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK106306 | 0.0 | AK106306.1 Oryza sativa Japonica Group cDNA clone:002-101-C10, full insert sequence. | |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP948 | 38 | 133 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 5e-99 | AP2 family protein | ||||



