 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN539799.1_FGP001 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1363aa MW: 149273 Da PI: 7.7349 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.7 | 2.1e-40 | 96 | 173 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C+adl++ak+yhrrhkvCe+h k++++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+q+
KN539799.1_FGP001 96 MCQVDDCRADLTNAKDYHRRHKVCEIHGKTTKALVGNQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQP 173
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 6.1E-34 | 89 | 158 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.949 | 94 | 171 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.49E-37 | 95 | 174 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.1E-29 | 97 | 170 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF49899 | 3.72E-27 | 1124 | 1329 | IPR013320 | Concanavalin A-like lectin/glucanase domain |
| Gene3D | G3DSA:2.60.120.200 | 5.4E-47 | 1126 | 1330 | IPR013320 | Concanavalin A-like lectin/glucanase domain |
| Pfam | PF00139 | 5.9E-24 | 1126 | 1241 | IPR001220 | Legume lectin domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0030246 | Molecular Function | carbohydrate binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1363 aa Download sequence Send to blast |
MWDWDSRALT AKPSSDALRV NAGLGLNLQL GLREDAATPM DVSPAATTVS SSPSPPASSA 60 PAQEPVVRPS KRVRSGSPGS ASGGGGEGGG GGSYPMCQVD DCRADLTNAK DYHRRHKVCE 120 IHGKTTKALV GNQMQRFCQQ CSRFHPLSEF DEGKRSCRRR LAGHNRRRRK TQPTDVASQL 180 LLPGNQENAA NRTQDIVNLI TVIARLQGSN VGKLPSIPPI PDKDNLVQII SKINSINNGN 240 SASKSPPSEA VDLNASHSQQ QDSVQRTTNG FEKQTNGLDK QTNGFDKQAD GFDKQAVPST 300 MDLLAVLSTA LATSNPDSNT SQSQGSSDCS GNNKSKSQST EPANVVNSHE KSIRVFSATR 360 KNGALERSPE MYKQPDQETP PYLSLRLFGS TEEDVPCKMD TANKYLSSES SNPLDERSPS 420 SSPPVTHKFF PIRSVDKDAR IADYGEDIAT VEVSTSRAWR APPLELFKDS ERPIENGSPP 480 NPAYQSCYTS TSCSDHSPST SNSDGQDRTG RIIFKLFGKE PSTIPGNLRG EIVNWLKHSP 540 NEMEGYIRPG CLVLSMYLSM PAIAWNELEE NLLQRVNTLV QGSDLDFWRK GRFLVRTDAQ 600 LVSYKDGATR LSKSWRTWNT PELTFVSPIA VVGGRKTSLI LKGRNLTIPG TQIHCTSTGK 660 YISKEVLCSA YPGTIYDDSG VETFDLPGEP HLILGRYFIE VENRFRGNSF PVIIANSSVC 720 QELRSLEAEL EGSQFVDGSS DDQAHDARRL KPKDEVLHFL NELGWLFQKA AASTSAEKSD 780 SSGLDLMYFS TARFRYLLLF SSERDWCSLT KTLLEILAKR SLASDELSQE TLEMLSEINL 840 LNRAVKRKSS HMARLLVQFV VVCPDDSKLY PFLPNVAGPG GLTPLHLAAS IEDAVDIVDA 900 LTDDPQQIGL SCWHSALDDD GQSPETYAKL RNNNAYNELV AQKLVDRKNN QVTIMVGKEE 960 IHMDQSGNVG EKNKSAIQAL QIRSCNQCAI LDAGLLRRPM HSRGLLARPY IHSMLAIAAV 1020 CVCVCVFMRA LLRFNSGRSF KWERLDFVIL LVDKTSPEIS VCNLQRALCL GKNKAWCVAN 1080 GFRCLLRELD AFAGHIHGDD DGANHLWKSN SSRLQRPSGE REPGGYDPDD NHVGLDVGTV 1140 ASNKTASLAG FNITIATNKT APANYTAWIE YDGAARRIAV YMGVRGAPRP ATPVLASPLD 1200 LSELVPERAY LGFTASTGVS FELNCILDWN LTIETFPADK KSKGWVVPVA VAVPVAAIAA 1260 AAFVVARMAR ARRSMERRRQ ERLEHTLTNL PGMPKEFAFE KLRKATKNFD ERLRLGKGGY 1320 GMVYKGVLPA AAVDDDDGRP AAARGGVGEE CIAKWSINLN SSD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 9e-32 | 90 | 170 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
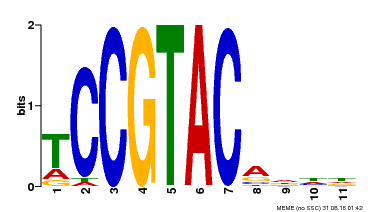
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN539799.1_FGP001 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK242070 | 0.0 | AK242070.1 Oryza sativa Japonica Group cDNA, clone: J075133L10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015649921.1 | 0.0 | squamosa promoter-binding-like protein 15 | ||||
| Swissprot | A2YX04 | 0.0 | SPL15_ORYSI; Squamosa promoter-binding-like protein 15 | ||||
| Swissprot | Q6Z8M8 | 0.0 | SPL15_ORYSJ; Squamosa promoter-binding-like protein 15 | ||||
| TrEMBL | A0A0E0IEE9 | 0.0 | A0A0E0IEE9_ORYNI; Uncharacterized protein | ||||
| TrEMBL | A0A0E0QL60 | 0.0 | A0A0E0QL60_ORYRU; Uncharacterized protein | ||||
| STRING | ORUFI08G22780.2 | 0.0 | (Oryza rufipogon) | ||||
| STRING | ONIVA08G22940.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5456 | 38 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||



