 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN540112.1_FGP003 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 504aa MW: 55293.9 Da PI: 5.6049 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 19.8 | 2.1e-06 | 265 | 286 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
C +Cgk F+r nL+ H+r H
KN540112.1_FGP003 265 FCLICGKGFKRDANLRMHMRGH 286
5*******************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 6.5E-5 | 262 | 290 | No hit | No description |
| PROSITE profile | PS50157 | 11.552 | 264 | 291 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 264 | 286 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.2E-5 | 266 | 290 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 266 | 286 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 68 | 313 | 346 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 22 | 351 | 373 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010044 | Biological Process | response to aluminum ion | ||||
| GO:0010447 | Biological Process | response to acidic pH | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 504 aa Download sequence Send to blast |
MASNATRNTD PDQQGVRFSS MDQPPCFARP GQSFPAFPPL FGVQSSSLYL PDDIEAKIGN 60 QFESNPSPNN PTMDWDPQAM LSNLSFLEQK IKQVKDIVQS MSNRESQVAG GSSEAQAKQQ 120 LVTADLTCII IQLISTAGSL LPSMKNPISS NPALRHLSNT LCAPMILGSN CNQRPSTNDE 180 ATIPDISKTP DYEELMNSLN TTQAESDEMM NCQNPCGGEG SEPIPMEDHD VKESDDGGER 240 ENLPPGSYVV LQLEKEEILA PHTHFCLICG KGFKRDANLR MHMRGHGDEY KTAAALAKPS 300 KDSSSESAPV TRYSCPYVGC KRNKEHKKFQ PLKTILCVKN HYKRSHCDKS YTCSRCNTKK 360 FSVIADLKTH EKHCGRDKWL CSCGTTFSRK DKLFGHVALF QGHTPALPMD DIKVTGASEQ 420 PQGSEAMNTM VGSAGYNFPG SSSDDIPNLD MKMADDPRYF SPLSFDPCFG GLDDFTRPGF 480 DISENPFSFL PSGSCSFGQQ NGDS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in aluminum tolerance. {ECO:0000250}. | |||||
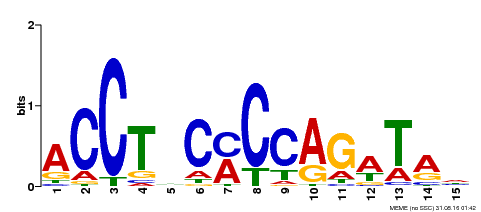
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00196 | ampDAP | Transfer from AT1G34370 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN540112.1_FGP003 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK072211 | 0.0 | AK072211.1 Oryza sativa Japonica Group cDNA clone:J013161E15, full insert sequence. | |||
| GenBank | AK072417 | 0.0 | AK072417.1 Oryza sativa Japonica Group cDNA clone:J023085P15, full insert sequence. | |||
| GenBank | AK100840 | 0.0 | AK100840.1 Oryza sativa Japonica Group cDNA clone:J023123F03, full insert sequence. | |||
| GenBank | AP003235 | 0.0 | AP003235.2 Oryza sativa Japonica Group genomic DNA, chromosome 1, PAC clone:P0039A07. | |||
| GenBank | AP003566 | 0.0 | AP003566.3 Oryza sativa Japonica Group genomic DNA, chromosome 1, BAC clone:OSJNBb0008G24. | |||
| GenBank | AP014957 | 0.0 | AP014957.1 Oryza sativa Japonica Group DNA, chromosome 1, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015641300.1 | 0.0 | zinc finger protein STOP1 homolog | ||||
| Refseq | XP_015641307.1 | 0.0 | zinc finger protein STOP1 homolog | ||||
| Swissprot | Q943I6 | 0.0 | STOP1_ORYSJ; Zinc finger protein STOP1 homolog | ||||
| TrEMBL | A0A0D3EWT7 | 0.0 | A0A0D3EWT7_9ORYZ; Uncharacterized protein | ||||
| TrEMBL | I1NTS4 | 0.0 | I1NTS4_ORYGL; Uncharacterized protein | ||||
| STRING | ORGLA01G0326400.1 | 0.0 | (Oryza glaberrima) | ||||
| STRING | OBART01G38810.1 | 0.0 | (Oryza barthii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1512 | 38 | 111 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34370.2 | 1e-158 | C2H2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



