 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN540599.1_FGP002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 366aa MW: 40105.3 Da PI: 8.1169 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 106.5 | 1.5e-33 | 29 | 83 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWtp+LHerFveav++LGG++kAtPk++l+lm++kgLtl+h+kSHLQkYRl
KN540599.1_FGP002 29 KPRLRWTPDLHERFVEAVTKLGGPDKATPKSVLRLMGMKGLTLYHLKSHLQKYRL 83
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.032 | 26 | 86 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-32 | 26 | 84 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.49E-16 | 29 | 86 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-24 | 29 | 84 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.3E-9 | 31 | 82 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 1.6E-24 | 128 | 172 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 366 aa Download sequence Send to blast |
MERAGYGVGV GVGGAGAVGA GVVLSRDPKP RLRWTPDLHE RFVEAVTKLG GPDKATPKSV 60 LRLMGMKGLT LYHLKSHLQK YRLGKQNKKD TGLEASRGAF AAHGISFASA APPTIPSAEN 120 NNAGETPLAD ALRYQIEVQR KLHEQLEVQK KLQMRIEAQG KYLQTILEKA QNNLSYDATG 180 TANLEATRTQ LTDFNLALSG FMNNVSQVCE QNNGELAKAI SEDNLRTTNL GFQLYHGIQE 240 SDDVKCSQDE GLLLLDLNIK GGGYDHLSSN AMRGGESGLK ISQHRSIKES FVVSFWSKHT 300 TSDDQSNQRQ AFCHFTTGEV VVGYLEEDVS GHAQRLDGPV GDLVEQNLRL VVLLQVDRRV 360 RRLILG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 8e-21 | 29 | 85 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 8e-21 | 29 | 85 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 8e-21 | 29 | 85 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 8e-21 | 29 | 85 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
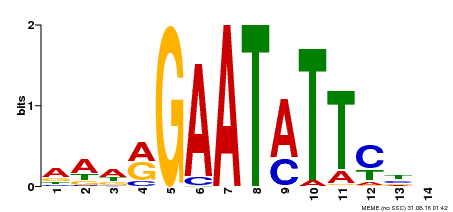
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN540599.1_FGP002 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK059448 | 0.0 | AK059448.1 Oryza sativa Japonica Group cDNA clone:001-028-A10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015623757.1 | 0.0 | myb family transcription factor PHL11 | ||||
| TrEMBL | Q6Z8D2 | 0.0 | Q6Z8D2_ORYSJ; Phosphate starvation response regulator-like | ||||
| STRING | OGLUM02G29720.1 | 0.0 | (Oryza glumipatula) | ||||
| STRING | ORUFI02G30700.1 | 0.0 | (Oryza rufipogon) | ||||
| STRING | OS02T0700300-01 | 0.0 | (Oryza sativa) | ||||
| STRING | ORGLA02G0249400.1 | 0.0 | (Oryza glaberrima) | ||||
| STRING | OBART02G29100.1 | 0.0 | (Oryza barthii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3940 | 34 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 9e-64 | G2-like family protein | ||||



