 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KN540616.1_FGP002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 307aa MW: 34259.4 Da PI: 6.6732 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.2 | 9.9e-14 | 84 | 134 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ g +k+++lg f+++ eAa+a+++a+ +++g
KN540616.1_FGP002 84 SKYRGVTLHK-CGRWEAR---M---GqllGKKYIYLGLFDSEIEAARAYDRAAIRFNG 134
89********.7******...5...334336**********99**********99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 4.65E-27 | 84 | 144 | No hit | No description |
| SuperFamily | SSF54171 | 3.79E-17 | 84 | 143 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.4E-9 | 84 | 134 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.3E-16 | 85 | 142 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.2E-33 | 85 | 148 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.107 | 85 | 142 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.8E-7 | 86 | 97 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 9.8E-7 | 124 | 144 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 307 aa Download sequence Send to blast |
MGVAHLGLRE AGVPGWFRYS SCRSENPLCR AYDRAAIKFR GLDADINFNL NDYEDDLKQM 60 RNWTKEEFVH ILRRQSTGFA RGSSKYRGVT LHKCGRWEAR MGQLLGKKYI YLGLFDSEIE 120 AARAYDRAAI RFNGREAVTN FDPSSYDGDV LPETDNEVVD GDIIDLNLRI SQPNVHELKN 180 DGTLTGFQLN CDSPEASSSV VTQPISPQWP VLPQGTSMSQ HPHLYASPCP GFFVNLREVP 240 MEKRPELGPQ SFPTSWSWQM QGSPLPLLPT AASSGFSTGT VADAARAPSS HPHPFPGHHQ 300 FYFPPTA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition (Probable). Together with IDS1, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes (PubMed:22003982). Prevents lemma and palea elongation as well as grain growth (PubMed:28066457). Regulates the transition from spikelet meristem to floral meristem, spikelet meristem determinancy and the floral organ development (PubMed:17144896). {ECO:0000250|UniProtKB:P47927, ECO:0000269|PubMed:17144896, ECO:0000269|PubMed:22003982, ECO:0000269|PubMed:28066457, ECO:0000305|PubMed:26631749}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
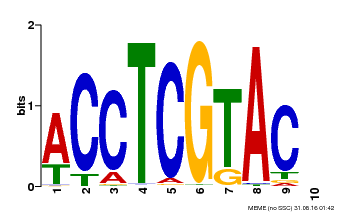
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KN540616.1_FGP002 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Target of miR172 microRNA mediated cleavage, particularly during floral organ development. {ECO:0000269|PubMed:20017947, ECO:0000269|PubMed:22003982, ECO:0000305|PubMed:26631749, ECO:0000305|PubMed:28066457}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CT831975 | 0.0 | CT831975.1 Oryza sativa (indica cultivar-group) cDNA clone:OSIGCRA106E13, full insert sequence. | |||
| GenBank | DQ374663 | 0.0 | DQ374663.1 Oryza sativa (japonica cultivar-group) supernumerary bract mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015647891.1 | 0.0 | ethylene-responsive transcription factor RAP2-7 isoform X1 | ||||
| Swissprot | Q8H443 | 0.0 | AP23_ORYSJ; APETALA2-like protein 3 | ||||
| TrEMBL | A0A0E0Q5Y6 | 0.0 | A0A0E0Q5Y6_ORYRU; Uncharacterized protein | ||||
| TrEMBL | I1Q994 | 0.0 | I1Q994_ORYGL; Uncharacterized protein | ||||
| STRING | ORUFI07G08180.2 | 0.0 | (Oryza rufipogon) | ||||
| STRING | ORGLA07G0068600.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2448 | 38 | 90 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 5e-72 | related to AP2.7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



