 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KXZ42308.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Gonium
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 405aa MW: 40567.6 Da PI: 6.8018 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.9 | 7.1e-20 | 143 | 193 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
++y+GVr+++ +g+++AeIrdp + r +lg+++taeeAa a+++a+++++g
KXZ42308.1 143 TKYRGVRQRP-WGKYAAEIRDPH----KgCRLWLGTYDTAEEAALAYDKAAREIRG 193
69********.**********73....359************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.4E-13 | 143 | 193 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.87E-31 | 143 | 202 | No hit | No description |
| SMART | SM00380 | 2.6E-38 | 144 | 207 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.3E-32 | 144 | 202 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.42 | 144 | 201 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.12E-22 | 144 | 201 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 2.9E-10 | 145 | 156 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.9E-10 | 167 | 183 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 405 aa Download sequence Send to blast |
MGGGAPPNFG RLSVNGPSSE GGAGHGSGGG AADGLAALPM PSRRPPLAPG GPAPLPAGLA 60 DHPAAGICST SYKSSGSHLG PYPGAPLLGA SPPSLGTSPA AGRASNSRRR NSSTNNVLSS 120 SLGAKASSSA PALSVKPDKN GATKYRGVRQ RPWGKYAAEI RDPHKGCRLW LGTYDTAEEA 180 ALAYDKAARE IRGPRAVVNF PNMPHSTASP HPDDESGQWD PLGESLGTSP LSSSFVTGTS 240 PLMGGSAPGR HAGGAGGRGD LHPGLMPNRV HKQRNAYGGF GSHKRDPEPI AEGDSDVDMD 300 DGDDGEDMDE GMAPGHGGGG GGRSRTRINV VTGVGSTRPP RQAAAGVAAA VAAARGGIDD 360 DFEPAPAPQP RGGGGGGGWG RQVSMDVEDE LADLADALLL LHESG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 6e-23 | 145 | 201 | 6 | 63 | ATERF1 |
| 3gcc_A | 6e-23 | 145 | 201 | 6 | 63 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
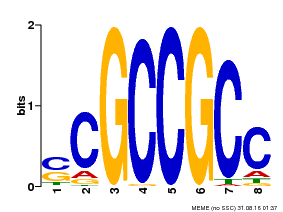
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_001699213.1 | 1e-107 | AP2-domain transcription factor | ||||
| TrEMBL | A0A150FXJ5 | 0.0 | A0A150FXJ5_GONPE; Uncharacterized protein | ||||
| STRING | EDO98853 | 1e-106 | (Chlamydomonas reinhardtii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP132 | 15 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G16770.1 | 4e-18 | ethylene-responsive element binding protein | ||||



