 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KXZ51980.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Gonium
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 1378aa MW: 135301 Da PI: 7.577 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 46.8 | 7e-15 | 56 | 100 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE+ ++++a k++G W++I +++ ++t+ q++s+ qk+
KXZ51980.1 56 RERWTEEEHAKFLEALKLYGRA-WRKIEEHVS-TKTAVQIRSHAQKF 100
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.39E-15 | 51 | 106 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.4 | 51 | 105 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-9 | 54 | 103 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.0E-15 | 54 | 103 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.2E-12 | 55 | 103 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.1E-12 | 56 | 99 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.28E-9 | 58 | 101 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010243 | Biological Process | response to organonitrogen compound | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0032922 | Biological Process | circadian regulation of gene expression | ||||
| GO:0042754 | Biological Process | negative regulation of circadian rhythm | ||||
| GO:0043496 | Biological Process | regulation of protein homodimerization activity | ||||
| GO:0043966 | Biological Process | histone H3 acetylation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1378 aa Download sequence Send to blast |
MEEVKGSKAG SAAVRAPAQP QPAGPASAPS SSAPLSDQQA CPKVKPRKPY IITKQRERWT 60 EEEHAKFLEA LKLYGRAWRK IEEHVSTKTA VQIRSHAQKF INKLERNKDT GSTEDGEGIS 120 IPPPRPKRKP SRPYPRKEPV ADNSGSGAAD ASGVNGGAGG GTVMGSQGTG PGTGQQQSKL 180 SMPVPPRSSH MLQAPMNALQ QLQLQMPLHL SAQQQQTLGP TVAAATAAAL KQQQERQQTS 240 HYSEVTDATV AAVAAAASAA AAAAAAAVVA AAGQQVQAHL QAHPPQGFPF FGLPPSLLAQ 300 ITLQNPALEA MPFPSGASSQ PGPPPLTSGG SGEATGHRPW ASSADAGPWM AVHPDRYHHL 360 HHHYHHYHHH GGDGRPQQPL AEATATTTDA ENSGPLTNGE LMNIAMGVGA GQESRGSDGN 420 SVVTEREGEQ GDGDGGDGDG EGDDTGPDAV AEDQAEQSNR ADTQWPDGGQ AAEPAGMTTS 480 QYWQMLGMID GGRPGHAGAA AAAGHAPREG HLHPHPHGLP HGHKAISNST RSTGSGGALR 540 AKGIAKAHTH KEPTAFARDQ HKKAAARTTS PCCSDMSNML RSHHHHHHHD PHSPSIQDVG 600 AKPPGRGSCD TNGSDSPLRA NRNDAEIREA QGSNPTNHNN GNGNGSSGNG SAGNGNGSGS 660 QGNGSQGNGH SGGNGHSGGN GHSGGNGHSG GSGGGGGSGG NGNGHSIKDS NNPTGNGHSS 720 GHGNGNGHSS ANGTSQHHHH HHHHHALPRA VAYDAGRYPS HHETQDRDGN GASGIGVRPP 780 SSEDEDIAVG EGAEAGVEAA VRPGDGGNAD GQELDNAGTT LDGDAGACQG AGGSTAPSDS 840 GQPGGGGSSR GVAGSGDKAK ESGVSAGAGG SGAGGSGSAD KPDRTGSGVV CGGGESGGGG 900 AAGRGVTASD SVIGRMVRIS SGNGAFRRPE GALGLASGPD RPVGQVAGSA FSIPPPLNPR 960 GGGAIGSSGA GARGSSAPTA PRSTGANKVQ TLLAQLQAHA SLAQYQAPAP GVPASFSQQA 1020 MHTATAAAQP FYATASLAAN VVHQGLLGFA APQAAAAGIP LPASAAVAAS AAAVAAAAAG 1080 KLAVPPATDS IPLAVALSQL AAQGGYGPRS AAEGLSDLPP VMPLSAGPAN SVAIAALGSI 1140 GAAASAVTGD AAGGSAGPAA YAQLVALMKA AAPQATRSEG AVVAAAAAAA LSPRAAQEAS 1200 EVGAAGGDGS SGFAGGAGMP RALSASAPLL AAAVLAAAQQ QQQCAAPRLL QAPRMRISSE 1260 GGLGGAAQQP VAAALASPRA VEASAADLRR RTVMVDGSNG AAAVAAAVRS AGAAASGGGR 1320 VTASGSGAAG AAAPVGMDRA EGMQEPAAAG SGKRSAGSRP SDTSGRPASP PRKRPHQT |
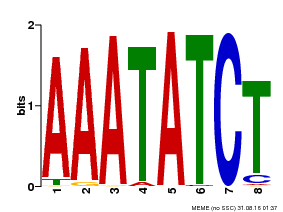
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00103 | PBM | Transfer from AT2G46830 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A150GRN4 | 0.0 | A0A150GRN4_GONPE; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP454 | 14 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46830.1 | 3e-29 | circadian clock associated 1 | ||||



