 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KXZ53699.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Volvocaceae; Gonium
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 788aa MW: 78119.8 Da PI: 4.7451 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 60.1 | 5e-19 | 417 | 466 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+++GVr+++ +g+++AeIrdp+++ r +lg+f+taeeAa+a+++a+++l+g
KXZ53699.1 417 RFRGVRQRP-WGKYAAEIRDPVKG---GRTWLGTFDTAEEAARAYDNAARALRG 466
69*******.**********7654...7************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.15E-26 | 417 | 474 | No hit | No description |
| PROSITE profile | PS51032 | 22.787 | 417 | 474 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.2E-35 | 417 | 480 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.8E-11 | 417 | 466 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.6E-30 | 417 | 475 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.97E-21 | 418 | 475 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 1.3E-9 | 418 | 429 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.3E-9 | 440 | 456 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 788 aa Download sequence Send to blast |
MDLSTGHPVG PSHLIKLSSV LEGSFESTAA LMRERQQRQQ QQQRQQQQQR SGPALSHPPP 60 PTQQQHAAEV RGGAGEARGG AAAGSVVGSA AWADELAAAA VNGGSRALNA TAVALAQAMR 120 NGGDEAAAAL TALHAFRRLD SKRLPLDERQ LMGWPGAAAA GLLLDREPSD DRGGASAGVR 180 VKRSDSVDRA DVGPVQLGGG RGGGGGERDG WEQQGRPGQT GGRAAGAAAG AGAGAGAAAG 240 HRAGGAVRQG GGGEAPFKLP IDDPFYRQQV EAMAAALGLS PQRATQLVLS HQQQLAAEDE 300 ARLAEAEAEL GEAAAMAAAA EAEAGAGPSG GRAASPAGGA TESVGATAGS GPRGAAGAEG 360 EGALAGSDAE ADEGDAGGEG RTDGDTSPNG AGGGRGQGGG GKARMGGIPA VGRGAVRFRG 420 VRQRPWGKYA AEIRDPVKGG RTWLGTFDTA EEAARAYDNA ARALRGPGAR VNFPLADGEV 480 PPPMTGVLPA VGGGGSGGGA GARGGDDGSG DEGPDGDDAP PSQPVMPAHE LRGLDVIALA 540 AELAAAEQAE LEQAEAERQR AAKRPRESSK AAAAAAGTRG SGHGTAAKRQ QRHKAAAAAA 600 APSAGPQAMD AEDEWIGPDS RRVNAPGREE SDGGQQHRQQ TEGLTPADLL EAALAAGDAL 660 DALAAEAEAE AHVAEGTEDA GGDSHRGPEG PEGGREFVGD LHSAVEAEGD TQAAAHDDRR 720 PLPPQEQQEG GRDPGQEEQR AAGSQGVGGM AAPEREQQAA CLSPRNDGAA LAAAAAAAVA 780 AGGTPRAC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 2e-20 | 417 | 474 | 14 | 72 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 392 | 400 | GGRGQGGGG |
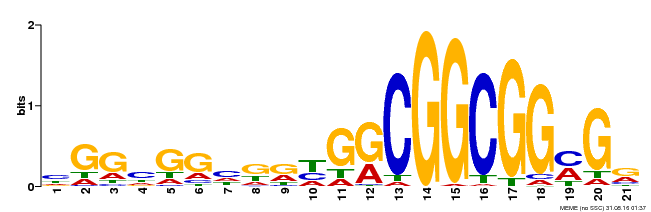
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00542 | DAP | Transfer from AT5G44210 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A150GV35 | 0.0 | A0A150GV35_GONPE; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP132 | 15 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G11140.1 | 2e-12 | cytokinin response factor 1 | ||||



