 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV15099.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 683aa MW: 73671.3 Da PI: 6.4698 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50 | 5.1e-16 | 429 | 475 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
KZV15099.1 429 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLDEAIEYLKTLQ 475
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 4.08E-17 | 421 | 479 | No hit | No description |
| SuperFamily | SSF47459 | 1.96E-20 | 422 | 485 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.228 | 425 | 474 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.3E-13 | 429 | 475 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.5E-20 | 429 | 483 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 431 | 480 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 683 aa Download sequence Send to blast |
MARGKLESGQ LKTSPTDNSS RMDGDFVELV WENGQPMMQG QSSRAARSLA SHSLQSNPPK 60 IRDAGVGHPT ISRIGKFGEV ETSLNDISVV VPTGELDLSQ DDEIDPWLNF PFDDALPHDY 120 CSEVFPETSG VTSSMHHISV PVDKSNSYDH TGRNLRNDGV GLKGLSCKAH ILSPWPPLQG 180 QTSDFSLGSQ FSDVISNSTS IYPDSVFGHS AQDRDLANDS ASKKVEWPSL KLPNNNLLNF 240 SHFSRSASLV KAKSPVHDGI PTTVSLGTEK VGGKDNVMVS SGPNLVKSAP DEPPSSSHKG 300 INLRVQPTVC EMGNSRPYMV EATKDLPAEN SVVSFVKNDN SITPSNCSSL KGVQDNDKNI 360 EPMVTSSSVG SDNCTDTVSC EKRPNYKRKL LDMEESECRS DGIQTGSVGV KKATSPGVSG 420 SKRSRAAEVH NLSERRRRDR INEKMRALQE LIPNCNKADK ASMLDEAIEY LKTLQLQVQI 480 MSMGAGLCMP PLMFPPGIQQ MHPTHVPHFS PMGMSMGMGF GTSMVDMNGG GTPRCPIFPL 540 PPMQAAQFSH PMSGPFNFPR VPGHSFPVYG YPSQGLHNSA PSVPLVALNR PPTDSAMGTG 600 KHNEVQSSSP NLNSEDPLTH NNSQRISNAE ASSSRNHGSN QLRTTTKVLD KPSIGQDNVQ 660 ATDVNFTESA SAAIDYTKEA GCD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 433 | 438 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
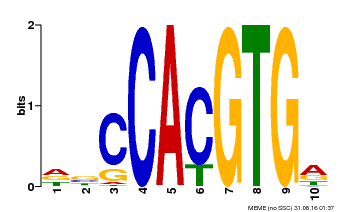
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV15099.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011087568.1 | 0.0 | transcription factor PIF3 isoform X3 | ||||
| TrEMBL | A0A2Z7A115 | 0.0 | A0A2Z7A115_9LAMI; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4965 | 24 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 5e-45 | phytochrome interacting factor 3 | ||||



