 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV16000.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 546aa MW: 60240.8 Da PI: 7.4735 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 56.2 | 6e-18 | 260 | 309 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f++Lr+++P++ ++K +Ka+ L +++eYI+ Lq
KZV16000.1 260 RSKHSATEQRRRSKINDRFQNLRDIVPNS----DQKRDKASFLLEVIEYIQFLQ 309
889*************************9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 5.1E-24 | 255 | 329 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 4.32E-19 | 256 | 324 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.206 | 258 | 308 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.1E-15 | 260 | 309 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.59E-14 | 260 | 312 | No hit | No description |
| SMART | SM00353 | 2.2E-13 | 264 | 314 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS51671 | 9.048 | 471 | 546 | IPR002912 | ACT domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016597 | Molecular Function | amino acid binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 546 aa Download sequence Send to blast |
MELPRPRGYE TEGRKATHDF LSLYSSSSPV QQDPTPQQGG YLETHDFLRP LERVGKNGTR 60 DESKDEMSVV DKLPLPAAEH LLPGGIGTYS ISYFNQRGLK PEGEGSLFMM QASSATRNDE 120 NSNSSSHTGS GFTLCDESAV KKGKTGKENF SAQRHVLQEA GLNMVGGQWP PASERASQSS 180 SNNKNNTATL GSFSSSQPAS SQKNHSFMSM IKSAKNDEEE CDDDDDKEFI MRKAPSFQPK 240 GNLSVKTETK TIDQKPNTPR SKHSATEQRR RSKINDRFQN LRDIVPNSDQ KRDKASFLLE 300 VIEYIQFLQE KVNKYESSYN VWNHETSKIV QRRNCHMGEG VIDHSQVFGS GPASALAARF 360 EENKIRVSPA MPINGQNLVE SDTSTATTFK EKAQQPELTS KAATLHLPMQ PGIFNLGRPS 420 AVASPLSPHQ ASDITKTTSW IPSHVLQSRS STDYWTRDKL KDHNLAIESG TISISSAYSQ 480 RLLSTLTLAL QESGIDLSQA SISVQIDLGK RSNGASSSTS TVKEDVPRDA SMEDEFSQAL 540 KRLKTI |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
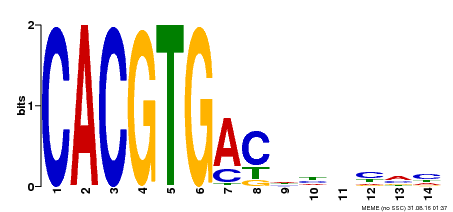
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV16000.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011078682.1 | 0.0 | transcription factor BIM1 isoform X1 | ||||
| TrEMBL | A0A2Z7AAL3 | 0.0 | A0A2Z7AAL3_9LAMI; Uncharacterized protein | ||||
| STRING | Migut.D01611.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3969 | 23 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 4e-95 | bHLH family protein | ||||



