 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV16313.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 337aa MW: 38234.5 Da PI: 9.0232 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 95.6 | 3.8e-30 | 65 | 118 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH +F++a+e+LGG+e+AtPk +l+lm+vkgL+++hvkSHLQ++R+
KZV16313.1 65 PRLRWTPELHICFLHAIERLGGQERATPKLVLQLMNVKGLSIAHVKSHLQMHRS 118
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-28 | 61 | 119 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 12.661 | 61 | 121 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.53E-15 | 62 | 119 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.0E-22 | 65 | 120 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 3.1E-8 | 66 | 117 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 337 aa Download sequence Send to blast |
MEDSKRSPQI QAEGDDDDHE ECKRDESVSD NKPTNGNSSS NSTVEESGKK ANRGSVRQYV 60 RSKTPRLRWT PELHICFLHA IERLGGQERA TPKLVLQLMN VKGLSIAHVK SHLQMHRSKK 120 IDDPNQVLSE QRLLLDAGDR HIYNLSQLPM LQSCNQTPIS SLRYRDYHWG RNSIYSPRSM 180 ASSYNNITRN GPYGSSSWKF LEKYKTNLET SRNQRSPQIQ LETSSATHNF APSRQDGGRE 240 HDDVHAHSPL PILKRTPKAP QDEMINTLKR KFSPEVNLDL NLSLKTTRVN EENAETSLNN 300 DEVDSTLTLS LFSTSSKHQV NGDKKDAKMT STLDLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 6e-17 | 66 | 120 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-17 | 66 | 120 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-17 | 66 | 120 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-17 | 66 | 120 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 6e-17 | 66 | 120 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
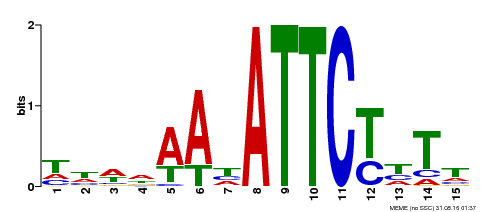
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV16313.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011076824.2 | 6e-94 | uncharacterized protein LOC105160975 | ||||
| TrEMBL | A0A2Z7A5C4 | 0.0 | A0A2Z7A5C4_9LAMI; Uncharacterized protein | ||||
| STRING | PGSC0003DMT400077217 | 5e-72 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA8573 | 21 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 3e-52 | G2-like family protein | ||||



