 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV19784.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 684aa MW: 76974.5 Da PI: 7.3305 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 140.6 | 1.4e-43 | 63 | 190 | 2 | 130 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqsslks 100
++ rk+++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+V++AL+reAGwvve+DGtty+++s + + s+ s++ s ++ +
KZV19784.1 63 AKSRKEREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVIAALAREAGWVVEADGTTYKQSSPAPPPPTPHSFSHLDSVQQSDHN-NAA 160
678******************************************************************98877755555555555555555453.344 PP
DUF822 101 salaspvesysaspksssfpspssldsisl 130
s pv s ++ ++sss + s ++si+
KZV19784.1 161 SVGVYPVRSVESPLSSSSLKNCSMMSSIEC 190
444566777776666666666666665554 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.0E-43 | 64 | 212 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene3D | G3DSA:3.20.20.80 | 3.8E-166 | 254 | 682 | IPR013781 | Glycoside hydrolase, catalytic domain |
| SuperFamily | SSF51445 | 1.73E-151 | 254 | 681 | IPR017853 | Glycoside hydrolase superfamily |
| Pfam | PF01373 | 1.2E-81 | 275 | 651 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 290 | 304 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 311 | 329 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 333 | 354 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 424 | 446 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 497 | 516 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 531 | 547 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 548 | 559 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 566 | 589 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.7E-51 | 604 | 626 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 684 aa Download sequence Send to blast |
MYSNTSPVNN RFVAADQDLQ LPQPQQKSPT ATTTYSHHPQ PQRRPRGFAA TFSTDSATST 60 IQAKSRKERE KEKERTKLRE RHRRAITSRM LAGLRQYGNF PLPARADMND VIAALAREAG 120 WVVEADGTTY KQSSPAPPPP TPHSFSHLDS VQQSDHNNAA SVGVYPVRSV ESPLSSSSLK 180 NCSMMSSIEC PPSGLRIDES LSPASLDSAV AERGVSLGNC AFASAVPSSE SLEPRKLVQE 240 VVYKEHGSGL SGTSFIPVYV KLSTGLVNNF CQLMDPEGIK QEIHHLKSLC VDGVVVDCWW 300 GIVEGWTPQK YEWAGYRELF NIIQEFGMKL KVVMAFHEHG GYGSGFISLP QWILEVGKIN 360 QDIFFTDREG RRNTECLSWG IDKKRVLRGR TGIEVYYDFM RSFRTEFDDF FIKGLITAVE 420 IGLGPSGELK YPSFSERMGW RYPGIGEFQC YDKYFLQDLG KAAKLRGHTV WARGPENAGY 480 YNSKPQETGF FSENGDYDSY YGRFFLQWYS QFLLDHADNV LTLARLAFEE IQIVVKIPAV 540 FWWYKTTSHA AELTAGYYNR ANQNGYSRVF DVLKKHAVTL NFLVSGPLAS SSEIDEASSD 600 HEGLSWQILS SAWDQGLSVA GQNAQPCLDK EGFTRLVETA KPRSHPDGRH FSFFTFQQHS 660 PLFQRTTCFA QIESFIKRMH GEHS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1v3i_A | 1e-123 | 254 | 684 | 10 | 446 | Beta-amylase |
| 1wdq_A | 1e-123 | 254 | 684 | 10 | 446 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
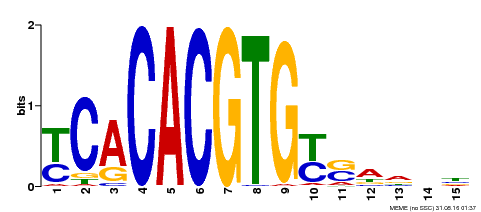
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV19784.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011082155.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A2Z7AKX7 | 0.0 | A0A2Z7AKX7_9LAMI; Beta-amylase | ||||
| STRING | Migut.A00486.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1607 | 24 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.2 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



