 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV29300.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 354aa MW: 39052.1 Da PI: 6.9957 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 91.5 | 6.5e-29 | 171 | 231 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqKe+ gs++prsYYrCt++ +Cp+kk+v+r ++dp ++e++Y+g+H+++
KZV29300.1 171 EDGYTWRKYGQKEIMGSTYPRSYYRCTHQkfyDCPAKKQVQRLNNDPFTFEVIYRGTHTCH 231
7****************************9999**************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF118290 | 1.31E-25 | 169 | 232 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 3.6E-27 | 169 | 232 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.6E-35 | 170 | 232 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.2E-26 | 171 | 231 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 21.172 | 171 | 233 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MEEALPLLYH GCKLAKDLEK SLQNSANNPE IILKSSEEII SIFSKIKEKI SLEQTSINVG 60 HDIGVGIQQW LKTSTVVLSG RDQPTPSHDP MHGHKISRMK GIQLPYEETG IGFKDIVQGS 120 GGPASDKQLL INAPDSTVTL PKQRKRKDSV SDKIVKRGAA PIMGNLELPP EDGYTWRKYG 180 QKEIMGSTYP RSYYRCTHQK FYDCPAKKQV QRLNNDPFTF EVIYRGTHTC HMSSTTPSTG 240 EPLPDQGPPS FTDTAAGIHL PQPFSSSLSA TTHWLSMQIF HDLVGVSAAV ASTNTAVARV 300 DSCGSAGPSA VRFHDYQLPV VDMADAMFNS NSSSSNSMDL IFSEHKWDLE EKKE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 4e-18 | 171 | 230 | 9 | 68 | OsWRKY45 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00304 | DAP | Transfer from AT2G40740 | Download |
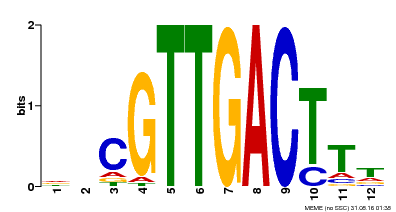
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV29300.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011076813.1 | 1e-103 | WRKY transcription factor 55 | ||||
| TrEMBL | A0A2Z7B5J7 | 0.0 | A0A2Z7B5J7_9LAMI; WRKY transcription factor 55 | ||||
| STRING | Migut.F00573.1.p | 1e-89 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA9280 | 18 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40740.1 | 4e-44 | WRKY DNA-binding protein 55 | ||||



