 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV33712.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 731aa MW: 80529.3 Da PI: 5.9944 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50 | 6.7e-16 | 24 | 68 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE+ ++++a k++G W +I +++g ++t+ q++s+ qk+
KZV33712.1 24 RERWTEEEHNKFLEALKLYGRA-WQRIEEHIG-TKTAVQIRSHAQKF 68
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.77E-16 | 18 | 72 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.989 | 19 | 73 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-8 | 21 | 68 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-16 | 22 | 71 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 4.7E-13 | 23 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.8E-13 | 24 | 67 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.45E-10 | 26 | 69 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010243 | Biological Process | response to organonitrogen compound | ||||
| GO:0042754 | Biological Process | negative regulation of circadian rhythm | ||||
| GO:0043496 | Biological Process | regulation of protein homodimerization activity | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 731 aa Download sequence Send to blast |
MDLYSSGEEL LLKTRKPYTI TKQRERWTEE EHNKFLEALK LYGRAWQRIE EHIGTKTAVQ 60 IRSHAQKFFT KLGGRARRGS VFLLVKGFNP WLEKESFVKG VPIGHTLNIN IPPPRPKRKP 120 SNPYPRKTST VVKDGKLSAP LSFLSQSQIT LDLEQEPLSE KSSDDGNNRK HVDREIENDS 180 AVFSLHETGA CANSSSLNKD SSASVAPINL CTFREYVPLS TDTSIQDETT QVFDAEQLRT 240 NQTCNKQLLQ EDGSFNTSNI GNSYLSHEKY CHDNRVDELK QSKNVDDLPT INDDVSQNCT 300 SHIPVHILEG SFGMNTQNVV LDTAHTDSRY QKMGVLGPPC LFMNPCGSTN PEHHSNASSS 360 SIHQPFPGFH PLSAAFQHQD DYQSFLHISS TFSNLLVSAL LQNPAAYAAA SFAATLWPCV 420 SGESSAGTPR GLQPKQMNTV PSMATIAAAT VAAATAWWAA HGLLPLCPPL DPGFTSTAPS 480 ASATPIDGNT RSENGEKKED NPDLALEDQQ LGQKCLAEFQ EQHSTLSSSC SEGIEGVKPN 540 CGSAVAETAP VADTVDKSNS RKQVDRSSCG SNTPSSSEVE AEGKEEKGVE DKEPKEDEEN 600 HPSGDPVNRR CRSLGSINDP WKEVSEEGRL AFQALFSREI LPQSFCHDLN FKRKKNSDME 660 STEGRPQLNL NDQGRLDDME EESTSKACRT GFKPYKRCSV EAKDTRMSTN CKDEEKGPKR 720 FRVGGIGKDS T |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00654 | PBM | Transfer from LOC_Os08g06110 | Download |
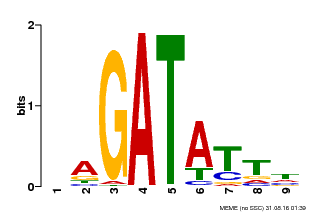
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV33712.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020553443.1 | 0.0 | protein LHY-like | ||||
| TrEMBL | A0A2Z7BH70 | 0.0 | A0A2Z7BH70_9LAMI; Protein LHY-like | ||||
| STRING | VIT_15s0048g02410.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA9710 | 13 | 15 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01060.3 | 5e-40 | MYB_related family protein | ||||



