 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV47133.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 435aa MW: 46767.6 Da PI: 5.8383 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 15.2 | 3.8e-05 | 273 | 296 | 32 | 55 |
..TTS-STCHHHHHHHHHHHHHHH CS
HLH 32 kapskKlsKaeiLekAveYIksLq 55
+++s++ +Ka++L +++eY+k+Lq
KZV47133.1 273 HNQSERTDKASMLDEVIEYVKQLQ 296
56999******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 8.765 | 242 | 295 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.9E-6 | 272 | 310 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.69E-4 | 273 | 300 | No hit | No description |
| SuperFamily | SSF47459 | 2.09E-7 | 274 | 324 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 435 aa Download sequence Send to blast |
MNQCVPSWDL DDNSALPGLD YEMAELTWKN GQLAMHGLGP PRVPPTKYTT WDKPRPGAGT 60 LESIVNQATR LPYPKYASDG SGDDVVPWFD HRKSMANPAA SASATMTMDA LVPCNKTNLI 120 NGNDNPVPSS QILESGTCAP GCSTRVGSCS GVAATLDGGV GGGNHVPRDV DFNSRETNGS 180 ASESATCDRD SRQLTLDTCE KDQMGAAWFT SPSTDSPENT SSGKEDSKST GEDHDSVRHI 240 RRQRDKGDID EIKKRGSGKS SVLTKRSRAA AIHNQSERTD KASMLDEVIE YVKQLQAQVQ 300 VMRMNMSHMN MMPLAAMQQH QHIQMSMMNP MGMGMGMGMG MGAGIMDINA MGRLGSGMLP 360 PPTAGIPPAP GSFMPVGAAW DNSGERLQGP TSVMTDPLSM FMAWQQSQIM TIDAYNRLAA 420 LYQQFQPPPG SSSKN |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
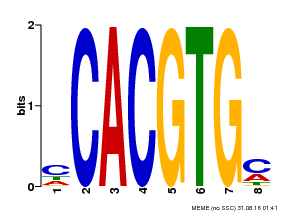
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV47133.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011093295.1 | 1e-136 | LOW QUALITY PROTEIN: transcription factor UNE10 | ||||
| TrEMBL | A0A2Z7CK54 | 0.0 | A0A2Z7CK54_9LAMI; Uncharacterized protein | ||||
| STRING | cassava4.1_007418m | 1e-107 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4367 | 21 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 2e-52 | bHLH family protein | ||||



