 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | KZV53783.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Gesneriaceae; Didymocarpoideae; Trichosporeae; Loxocarpinae; Dorcoceras
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 480aa MW: 52859.8 Da PI: 6.4083 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 34.6 | 4.8e-11 | 202 | 258 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + rk + g ++ +e+Aa+a++ a++k++g
KZV53783.1 202 SIYRGVTRHRWTGRYEAHLWDnSCR-RegqARKGRQ-GGYDKEEKAARAYDLAALKYWG 258
57*******************4444.2344336655.779999**************98 PP
| |||||||
| 2 | AP2 | 49.9 | 7.8e-16 | 301 | 352 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
KZV53783.1 301 SIYRGVTRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 352
57****************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.0E-8 | 202 | 258 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.14E-20 | 202 | 268 | No hit | No description |
| SuperFamily | SSF54171 | 4.58E-15 | 202 | 268 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 1.1E-12 | 203 | 267 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.1E-22 | 203 | 272 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.583 | 203 | 266 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.0E-6 | 204 | 215 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 6.16E-12 | 301 | 362 | No hit | No description |
| Pfam | PF00847 | 4.1E-10 | 301 | 352 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.19E-18 | 301 | 362 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.4E-18 | 302 | 361 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.151 | 302 | 360 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 9.3E-32 | 302 | 366 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.0E-6 | 342 | 362 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 480 aa Download sequence Send to blast |
MAPENNWLSF SLQSSVDQML NSSSHSADSH HFYSFSDNNY YANVENHQLV SYMDSSQVQP 60 PPKLEDFFAG DSTETQDSSL THIYDHSHHT GGGASAAAYF FSHGGEQQEL NNITGFQAFS 120 GNSGSEVDDT SFGGQIQSPV ESGNELAAYS QFPFTATTNA LSLGVSSKSN PLGAENNSCK 180 AIVSVDSDST KKISDTFGQR TSIYRGVTRH RWTGRYEAHL WDNSCRREGQ ARKGRQGGYD 240 KEEKAARAYD LAALKYWGST ATTNFPISNY TKEMDDMKNE TKQEFIASLR RKSSGFSRGA 300 SIYRGVTRHH QQGRWQARIG RVAGNKDLYL GTFATEEEAA EAYDIAAIKF RGVNAVTNFE 360 MSRYNVEAIS KSSLPIGGAA KRLKLSLESE ETQPISNILQ TQSCSSSSSI SFASTPTVSN 420 IPCGIPFDTS VPFYHHNFFQ QLQTSNSVAT DSSARVSSMA TPMPFMSSQP EFFIWPHQSL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00589 | DAP | Transfer from AT5G65510 | Download |
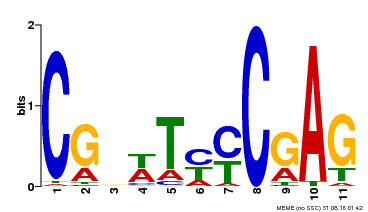
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | KZV53783.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011097469.1 | 0.0 | AP2-like ethylene-responsive transcription factor AIL6 isoform X1 | ||||
| Swissprot | Q52QU2 | 1e-143 | AIL6_ARATH; AP2-like ethylene-responsive transcription factor AIL6 | ||||
| TrEMBL | A0A2G9GKN6 | 0.0 | A0A2G9GKN6_9LAMI; Uncharacterized protein | ||||
| STRING | XP_009603183.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2580 | 24 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65510.1 | 1e-138 | AINTEGUMENTA-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



