 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0001s0109.5.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 396aa MW: 42764.6 Da PI: 6.644 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 68.7 | 9e-22 | 251 | 313 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e++lk errkq+NRe+ArrsR+RK+ae+eeL+ kv++L aeN++Lk+e+++l k+++klk e+
Kaladp0001s0109.5.p 251 ERQLKQERRKQSNRESARRSRLRKQAETEELAAKVDVLMAENMTLKSEINQLIKNSEKLKFEN 313
799********************************************************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 2.7E-26 | 1 | 92 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Gene3D | G3DSA:1.20.5.170 | 9.0E-15 | 249 | 311 | No hit | No description |
| Pfam | PF00170 | 1.1E-19 | 251 | 313 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 9.9E-19 | 251 | 315 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.449 | 253 | 316 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 7.5E-11 | 254 | 310 | No hit | No description |
| CDD | cd14702 | 4.40E-19 | 256 | 302 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 258 | 273 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 396 aa Download sequence Send to blast |
MGNNEESKSL KPDTTPLDQN GIHVYPPPDW AAMQAFYGPR IPLPQYYNSA YPAGHPPHPF 60 MWGPPQAMMP SFSAPYAAIY PQGGVYAHPG VPLGSPKPGF GALSTPSTTE ASMMQAATAL 120 TIQIPGKTSG NTTGGSRDSE SGDSGGSSGG SREKTSADEQ SHRKRKCEQS LDIEKTVKIE 180 IQANSIAPVD TNLESSMVAG IAVGPAANGK LENVTVLDLD MKPNHIVNSK MSFDAQQSGS 240 KLSSEACFQS ERQLKQERRK QSNRESARRS RLRKQAETEE LAAKVDVLMA ENMTLKSEIN 300 QLIKNSEKLK FENARMMEKL KNAKTEETER IGFNNADDSR SQTLSTENLL SRVNNSGSVV 360 TPEEEGEMHG NNNSNPTKLR QLLDASPRAD AVAAS* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 267 | 273 | RRSRLRK |
| 2 | 267 | 274 | RRSRLRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00318 | DAP | Transfer from AT2G46270 | Download |
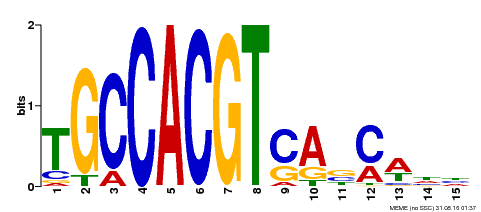
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021613119.1 | 1e-112 | common plant regulatory factor 1-like isoform X2 | ||||
| TrEMBL | A0A2P5E8F9 | 1e-112 | A0A2P5E8F9_TREOI; Basic-leucine zipper transcription factor | ||||
| STRING | cassava4.1_008459m | 1e-112 | (Manihot esculenta) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46270.2 | 7e-38 | G-box binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0001s0109.5.p |



