 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0008s0410.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 712aa MW: 78101.3 Da PI: 8.4742 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 74.8 | 1e-23 | 108 | 209 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f k+lt+sd+++ g +++p+ +ae++ ++ + t++ +d +g+ W++++iyr++++r++lt+GW+ Fv++++L +gD++vF
Kaladp0008s0410.1.p 108 FAKTLTQSDANNGGGFSVPRYCAETIfprldytADPPVQ--TVIAKDVHGEIWKFRHIYRGTPRRHLLTTGWSTFVNQKKLVAGDSIVFL 195
89****************************997556555..8************************************************ PP
E-SSSEE..EEEEE-S CS
B3 84 ldgrsefelvvkvfrk 99
++ ++el+v+++r+
Kaladp0008s0410.1.p 196 --RTGSGELCVGIRRA 209
..4479999*****97 PP
| |||||||
| 2 | Auxin_resp | 114.2 | 1.2e-37 | 278 | 361 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrl.sGtvvgvsdldpvrWpnSkWrsLk 83
aa+ a+++ +FevvY+Pra+t+eF+v ++ v++a+++++++GmRfkm fetedss++++ +Gtv++v+ +dp+rWpnS+Wr+L+
Kaladp0008s0410.1.p 278 AATLATNGRPFEVVYYPRACTPEFCVAASAVRAAMRIQWCPGMRFKMPFETEDSSRISWfMGTVSSVQVADPLRWPNSPWRLLQ 361
68899*****************************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 8.5E-38 | 99 | 217 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.07E-28 | 103 | 227 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.70E-21 | 107 | 208 | No hit | No description |
| SMART | SM01019 | 6.5E-22 | 108 | 210 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 9.8E-22 | 108 | 209 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.155 | 108 | 210 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 2.7E-32 | 278 | 361 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 15.715 | 597 | 677 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007389 | Biological Process | pattern specification process | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0048829 | Biological Process | root cap development | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 712 aa Download sequence Send to blast |
MERSLDSQLW HACAGGMVQM PKVNSKVYYF PQGHAEHAQA GLDFALSPKA GPLVPCRVAS 60 VQFLADPETD EVYAKLRLVP VGNRELEFGV EDLGVCRGSE SVDKPSSFAK TLTQSDANNG 120 GGFSVPRYCA ETIFPRLDYT ADPPVQTVIA KDVHGEIWKF RHIYRGTPRR HLLTTGWSTF 180 VNQKKLVAGD SIVFLRTGSG ELCVGIRRAK RGMESESGSG IRSQSGWSPS PGHFISPYGG 240 GGYSMFLRDD ENGFLRNGCG EGGLKTNKKV KGESVIEAAT LATNGRPFEV VYYPRACTPE 300 FCVAASAVRA AMRIQWCPGM RFKMPFETED SSRISWFMGT VSSVQVADPL RWPNSPWRLL 360 QVTWDEPDLL QNVNRVSPWL VELVSNTPAM NLPPFSSPRK KLRFPPQPDF PLDGQLSLSS 420 FSGNNLGPRS SFCCLIDNTP PGIQGARHAQ FGSSLSDIHF NSKLRSWLFP QSQQRLDQFP 480 RLFNNIISSS AKGNESESVS CLLSIGNSDH NVEKPKDGKK QPFVLFGQPI LTEQEISLRS 540 ESTLQALARN HSSDDHLERG KVHTSGLELR RSLSDTSSHP GFSRLFNFAS DELSMASGRC 600 EVFMDNDGVG RTLDLSPIQS YQQLCLKLAD LFGIGNPNAL NHALYRDATG AIKRIGDEPF 660 SDFRKSAKKL IITVDLGKKN NGREQYLSGP GKAENGLCSS KAAAGHLSIL A* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 6e-81 | 5 | 382 | 51 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
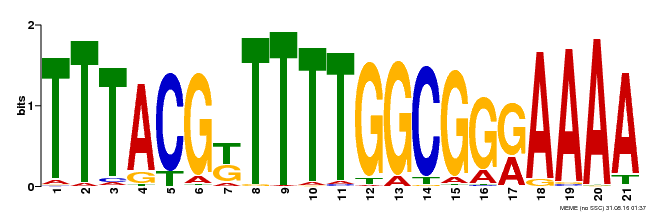
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00461 | DAP | Transfer from AT4G30080 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023882698.1 | 0.0 | auxin response factor 18 | ||||
| Refseq | XP_023882705.1 | 0.0 | auxin response factor 18 | ||||
| Swissprot | Q653H7 | 0.0 | ARFR_ORYSJ; Auxin response factor 18 | ||||
| TrEMBL | A0A438DTG8 | 0.0 | A0A438DTG8_VITVI; Auxin response factor | ||||
| STRING | VIT_06s0004g02750.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G30080.1 | 0.0 | auxin response factor 16 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0008s0410.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



