 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0011s0508.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 529aa MW: 58931.7 Da PI: 7.656 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 31.7 | 3.4e-10 | 231 | 273 | 2 | 41 |
XXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHH...HHHHHHHH CS
bZIP_1 2 kelkrerrkqkNReAArrsRqRKkaeieeLee...kvkeLeae 41
++ k rr+++NReAAr+sR+RKka++++Le + ++Le+e
Kaladp0011s0508.1.p 231 RDAKTLRRLAQNREAARKSRLRKKAYVQQLESsriRLAQLEQE 273
678999************************9633356666665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 7.1E-8 | 227 | 279 | No hit | No description |
| SMART | SM00338 | 0.0012 | 230 | 283 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.4E-7 | 231 | 273 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 8.921 | 232 | 276 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.34E-7 | 234 | 272 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 237 | 252 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 5.1E-31 | 327 | 402 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 529 aa Download sequence Send to blast |
MASSAHNNDH PSRQQQEAKQ HGPHHQFSSS GMMMMMMQPL HPSSSSSPSL HPKLICAKDD 60 RGGAAYGLGE LDEALFLYLN RQPQHEVAEV HHQKQKQQQQ QLELEHHHHS IIDLQQQITH 120 DGGMRPTLNI FPSQPMHVEP PSATNQVNNK SNETDPRGLV VVVSGAAQQG QVVSGGSSSK 180 MVSAAEPIST DILPNSTLIL SNTNQGKRTS SNSKRGPASS SELQITRSQT RDAKTLRRLA 240 QNREAARKSR LRKKAYVQQL ESSRIRLAQL EQEIQRARSQ GAHFGGGLIL GAEQHLGLSG 300 GINNFTTDAA IFDMEYARWL EEHHRLMCEL RAAIEEHLSE NELQIYVDNC LAHFDEIMNI 360 KSIMAKTDVF HLISGSWKSP AERCFMWIGG FRPSQIIKVV MSKIEGLTDE QAVGICGLQR 420 SMEQAEAALS HGLEELNQSL SDAIVYDSLT SPPRDMTNYM GQMAMAASKL SVLEGFVRQA 480 DNLRYQAIHR LHQILMTRQT ARGLLLIAEY FHRLRALSSL WLARPPLE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to as1-like elements (5'-TGACGTAAgggaTGACGCA-3') in promoters of target genes. Regulates transcription in response to plant signaling molecules salicylic acid (SA), methyl jasmonate (MJ) and auxin (2,4D) only in leaves. Prevents lateral branching and may repress defense signaling. {ECO:0000269|PubMed:12777042}. | |||||
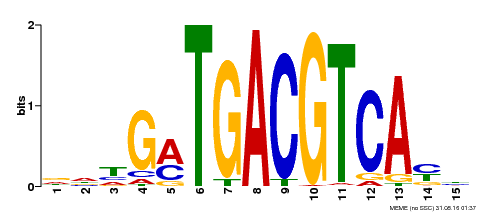
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00491 | DAP | Transfer from AT5G06839 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018503110.1 | 0.0 | PREDICTED: transcription factor TGA2.3-like | ||||
| Swissprot | Q52MZ2 | 1e-177 | TGA10_TOBAC; bZIP transcription factor TGA10 | ||||
| TrEMBL | A0A314YV58 | 0.0 | A0A314YV58_PRUYE; BZIP transcription factor TGA10 isoform X1 | ||||
| TrEMBL | A0A498HYB5 | 0.0 | A0A498HYB5_MALDO; Uncharacterized protein | ||||
| STRING | XP_009357109.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06839.3 | 1e-148 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0011s0508.1.p |



