 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0011s1015.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1291aa MW: 143280 Da PI: 8.9324 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.9 | 0.00016 | 1198 | 1220 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk+F ++ +L +H r+H
Kaladp0011s1015.1.p 1198 CPvtGCGKTFFSHKYLLQHRRVH 1220
9999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 10.9 | 0.0014 | 1256 | 1282 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y C Cg++F+ s++ rH r+ H
Kaladp0011s1015.1.p 1256 YMCAepGCGQTFRFVSDFSRHKRKtgH 1282
789999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.7E-17 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.478 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 8.7E-15 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 1.3E-46 | 195 | 364 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.078 | 198 | 364 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 9.06E-26 | 213 | 380 | No hit | No description |
| Pfam | PF02373 | 2.8E-37 | 228 | 347 | IPR003347 | JmjC domain |
| SMART | SM00355 | 26 | 1173 | 1195 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.23 | 1196 | 1220 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.05 | 1196 | 1225 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.9E-5 | 1198 | 1219 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1198 | 1220 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 8.09E-6 | 1198 | 1232 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.7E-8 | 1220 | 1248 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0014 | 1226 | 1250 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.471 | 1226 | 1255 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1228 | 1250 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.05E-9 | 1236 | 1281 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.3E-9 | 1249 | 1279 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 1.1 | 1256 | 1282 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.845 | 1256 | 1287 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1258 | 1282 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1291 aa Download sequence Send to blast |
MSGKQQQAEV FQWLRALPVA PEYRPTLDEF QDPIAYIFKI EKEASRYGIC KIVPPLPPAP 60 RKTVVANFNK SVPNGSGARP TFTTRQQQIG FCPRRGRPVQ RPVWQSGEFY TFQEFEAKAR 120 AFERSYLKKC GKKAAVAAAA AAGPGLSALE IETLYWKASA DKPFSVEYAN DMPGSAFAAA 180 SGTGRRPREG GEGMTVGDTA WNMKGVARST MSLLRFMKED IAGVTSPMVY VAMLFSWFAW 240 HVEDHDLHSL NYLHTGAGKT WYGVPKEAAA GFEEVVRVHG YGGEINPLVT FATLGEKTTV 300 MSPEVFVKAG IPCCRLVQNP GEFVVTFPRA YHSGFSHGFN CGEASNIATP YWLAVAKEAA 360 VRRASINYPP MVSHFQLLYD LAFALCSSLP MGVHSQPRSS RLKQKRKGVG ETVVKQLFVE 420 DIKESNRLLQ LLGEGLPVVL RPLSSAVFSI YSKSRIQCLL KSRSSSHLGL CYQKETVTSI 480 RNNLATSDIN SGTNGRLPTV IGLPAVGSKF CSTFDGNSRS LPNESTEPQL STSSRWVTDF 540 ENSNAIQDNG ASDKKLFSCV TCGILCFSCV AIIQPAEVAA RYLMASDCSF FSDSENAAKK 600 TSEELADSGR GVNTLDHTYS SGWMGRRAHN GTSNHIEEEY QSDEANSDRE EQPRTTALDL 660 LASAYGNSSD SENDTRLDSD PVYADGPNFS KISSDSDFEC NDSDIRDPLQ ENDFRWESSP 720 RLHSEGETTR FHIADSDGEN HYRRAFQNRN STYDSPSFNL DKLASMGSNL ASLQSDKDSL 780 RMHVFCLEHA IEVKKKLKQI GGVNMLLLCH PDYPRVEAEA KSVQEELGLD HCWNDTAFRE 840 SSEDDERCMA AALESAEDVS SNGDWTVKLG INLFYSASLS RSPLYSKQMP YNSIVYKAFG 900 RVPPLEAPAK SDGHTKGNSR KKKIVLAGKW CGKPWMSNQV HPLLLPPPPA LEETQQEHLH 960 AHASAKTDAA DSDSIQPVKS SKARRDDSSA TTSTPLAGSK RQIVVYSRSN KKTKRAETGL 1020 SVSASDGGDA DAGENLSNQQ AKLHVRSKHK IEGNKKPLEE ETRILYYSSK HVAKGDLKST 1080 KETTSLRDLY KPNFKPFGAD LDDKREGGLS TRLRRRNTKP ELEHEGKVLV KKERKPNKTP 1140 ANSKIEKVDA KPRKSKVSEA ARSRKKKDEE GEFQCDKEGC TMSFSLKQEL ALHKRDICPV 1200 TGCGKTFFSH KYLLQHRRVH EDARPLKCPW KGCKMTFKWA WARTEHIRVH TGVRPYMCAE 1260 PGCGQTFRFV SDFSRHKRKT GHSVKRNIKC * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 1e-75 | 7 | 388 | 2 | 356 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 1e-75 | 7 | 388 | 2 | 356 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
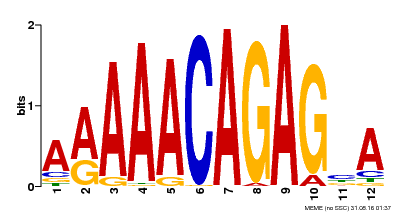
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010663055.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | F6GTM2 | 0.0 | F6GTM2_VITVI; Uncharacterized protein | ||||
| STRING | VIT_17s0000g09980.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0011s1015.1.p |



