 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0016s0134.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 270aa MW: 29848.6 Da PI: 7.4428 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.8 | 4.5e-17 | 15 | 62 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W+ eEd+ l+ +v q+G g+W++ ++ g+ R++k+c++rw +yl
Kaladp0016s0134.1.p 15 KGPWSLEEDQVLISYVSQHGHGRWRSLPKLAGLLRCGKSCRLRWTNYL 62
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 45.2 | 2.1e-14 | 69 | 113 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
++T+eE++l +++ lG++ W++Ia+ ++ gRt++++k+ w+++l
Kaladp0016s0134.1.p 69 APFTEEEEKLVTQLHGILGNR-WTAIAAQLP-GRTDNEIKNLWNTHL 113
68*******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.3E-24 | 6 | 65 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 23.862 | 10 | 66 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.38E-30 | 12 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.5E-13 | 14 | 64 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.3E-15 | 15 | 62 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.64E-10 | 17 | 62 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-24 | 66 | 117 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.786 | 67 | 117 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.1E-13 | 67 | 115 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.8E-13 | 69 | 113 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.28E-10 | 70 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 270 aa Download sequence Send to blast |
MGKKRPCCEK NGLKKGPWSL EEDQVLISYV SQHGHGRWRS LPKLAGLLRC GKSCRLRWTN 60 YLRPDIKRAP FTEEEEKLVT QLHGILGNRW TAIAAQLPGR TDNEIKNLWN THLRKRLLCM 120 GIDPLTHEPF ATSNGPALAP AAQWESARLE AEARLSRPRE SLQPGASSDH FLRLWNSEVG 180 EAFRLRARGA YVSSTSSIKG CCDSDMASTP AAKIGKGETC TSPTAASSSS DELDDSSDSA 240 LQLLTDYAFD DDMSFLADHI SDYYSFPSM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 9e-31 | 13 | 117 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may play a role in flower development by repressing ANT (PubMed:19232308). Regulates the transition of meristem identity from vegetative growth to flowering. Acts downstream of LFY and upstream of AP1. Directly activates AP1 to promote floral fate. Together with LFY and AP1 may constitute a regulatory network that contributes to an abrupt and robust meristem identity transition (PubMed:21750030). {ECO:0000269|PubMed:19232308, ECO:0000269|PubMed:21750030}. | |||||
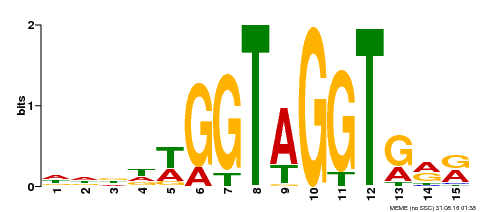
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00114 | ampDAP | Transfer from AT3G61250 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010661750.1 | 1e-111 | PREDICTED: myb-related protein 308 | ||||
| Refseq | XP_010661751.1 | 1e-111 | PREDICTED: myb-related protein 308 | ||||
| Swissprot | Q9M2D9 | 1e-103 | MYB17_ARATH; Transcription factor MYB17 | ||||
| TrEMBL | D7UC60 | 1e-109 | D7UC60_VITVI; Uncharacterized protein | ||||
| STRING | VIT_15s0046g03190.t01 | 1e-110 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61250.1 | 4e-96 | myb domain protein 17 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0016s0134.1.p |



