 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0022s0244.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 345aa MW: 39348.1 Da PI: 6.5164 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 84.1 | 1.8e-26 | 34 | 113 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
W ++e+++Li++rre+++ ++++k++k+lWe++s+kmr++gf rsp++C++kw+nl k+ykk+k++ +++y+++le
Kaladp0022s0244.1.p 34 WVQEETRCLINFRREVDSLFNTSKSNKHLWEQISIKMRDKGFDRSPTMCTDKWRNLLKEYKKLKHHHATT-----AKISYYKDLE 113
******************************************************************9973.....4577999997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 7.2E-4 | 26 | 92 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 8.177 | 26 | 90 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 1.2E-4 | 30 | 92 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 5.00E-27 | 32 | 97 | No hit | No description |
| Pfam | PF13837 | 2.5E-19 | 33 | 114 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 345 aa Download sequence Send to blast |
MMMEGAVSLA PAHQLLLGES SGDDKAPKKR AETWVQEETR CLINFRREVD SLFNTSKSNK 60 HLWEQISIKM RDKGFDRSPT MCTDKWRNLL KEYKKLKHHH ATTAKISYYK DLEDLMRDRT 120 SHTNAPLHSN KLLDSSSYIH FSHKAIEDAT LAFGHSEAGG HAKVNPESRL DHDSDPLAIN 180 TGETVVPNGV PPWNWRETAG SGEEQPTYDG RVISVKWGEY TRRIGIDGTT DAIKEAVKSA 240 FRLRTKRAFW LEDEYQIIRS IDRDMPLGNY TLHLDEGISI KVCLFDDAGR ITEEKTLYTE 300 DDFREYLQRR GLTGLRELNG YRSIDTLDDL GTGEVYHGVR MIGE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 2e-40 | 28 | 111 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00666 | PBM | Transfer from GSVIVG01007522001 | Download |
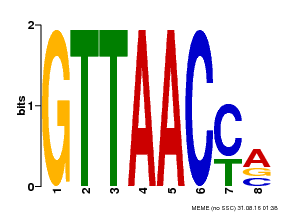
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010250844.1 | 1e-166 | PREDICTED: trihelix transcription factor GT-1 | ||||
| Swissprot | Q9FX53 | 1e-140 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A1U7ZBE5 | 1e-165 | A0A1U7ZBE5_NELNU; trihelix transcription factor GT-1 | ||||
| STRING | XP_010250844.1 | 1e-166 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 1e-131 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0022s0244.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



