 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0034s0224.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 746aa MW: 80718.1 Da PI: 6.3879 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 49.6 | 7.8e-16 | 447 | 485 | 2 | 40 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
++k+CnCkkskClk+YCeCfaag++C +C C++C+Nk
Kaladp0034s0224.1.p 447 SCKRCNCKKSKCLKLYCECFAAGVYCIGTCACQNCFNKP 485
689**********************************96 PP
| |||||||
| 2 | TCR | 49.4 | 9.4e-16 | 532 | 570 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
Kaladp0034s0224.1.p 532 RHKRGCNCKKSSCLKKYCECYQGGVGCSVNCRCESCKNV 570
589***********************************6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 3.4E-16 | 446 | 487 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 36.889 | 447 | 572 | IPR005172 | CRC domain |
| Pfam | PF03638 | 2.6E-11 | 449 | 484 | IPR005172 | CRC domain |
| SMART | SM01114 | 2.5E-15 | 532 | 573 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 2.1E-11 | 534 | 569 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 746 aa Download sequence Send to blast |
MDTPERNQIG SSLSKHEDSP VFNYLSNLSP IKPMKSAPIT QTFTTLNFSS LPSIFTSPHV 60 SSHKESRFLL RRHNTPDKSQ SEFSSYIDVS SNKENDVFVD VNQLHNCSSE LQKVSDTLDS 120 VGRAPLEPLH EESDLSIDLS HNLKNDCGSP DSEGKAARDE ASQSAEAATS SVMLDQGPSI 180 SLPAVHETPS GWESLISDGA DLLMFDSPNI AAGLKELFKS PPVEHMGSSF ASQFHQDTPD 240 FLKSDPAFLV DVGELQNETY HHAETEVIDC ARDDFPSTMT GCMTSHPSEN VDDESMMSLH 300 RGVRRRCLVF DTKGAQRKSL IGASSSNPSM CMNDNKKRDA SKIGGSSALC ILPGIGLHLN 360 ALATSGDRGG GQVSVSSSAT TFQSPATAHE SVLDVFSGAS SQENADSGII QCSEDACLAP 420 GLVSLDDINQ NSPRKKRRKP ESSGDESCKR CNCKKSKCLK LYCECFAAGV YCIGTCACQN 480 CFNKPVHEDT VLATRKQIES RNPLAFAPKV IRTSDPLPDI KDEVMNTPAS ARHKRGCNCK 540 KSSCLKKYCE CYQGGVGCSV NCRCESCKNV FGTKDGSAPV AVATQDGDEE EETEKVEMNN 600 KGLQSVMPHN NEEHNPNTFL ATPLRLTRMS MQLPFSSKGK PPRSSFLAVD SASGLQASQK 660 FSKPNYVNPP PKFQRNYPAG QDETEMPEML RNNGSPISAV KTSSPNSKRV SSPHSNFGSS 720 PAPRSTRKLI LQSIPSFPSL TQPAD* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 1e-16 | 451 | 570 | 14 | 121 | Protein lin-54 homolog |
| 5fd3_B | 1e-16 | 451 | 570 | 14 | 121 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 432 | 439 | PRKKRRKP |
| 2 | 434 | 438 | KKRRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
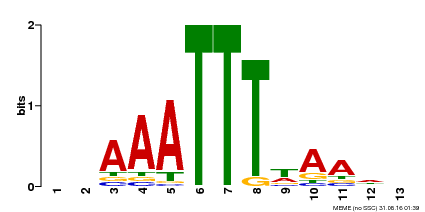
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018818657.1 | 0.0 | PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X3 | ||||
| TrEMBL | A0A2I4EGY1 | 0.0 | A0A2I4EGY1_JUGRE; protein tesmin/TSO1-like CXC 2 isoform X3 | ||||
| STRING | XP_008224266.1 | 0.0 | (Prunus mume) | ||||
| STRING | VIT_05s0020g03880.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 1e-116 | TESMIN/TSO1-like CXC 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0034s0224.1.p |



