 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0036s0258.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 292aa MW: 32482.9 Da PI: 8.7685 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 23.7 | 1.2e-07 | 40 | 83 | 3 | 44 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrw 44
+W++eE + + +a + + + +W ++a++++ g+t +++ +
Kaladp0036s0258.1.p 40 KWSQEENKMFENALAVYDRDspdRWQKVAAMIP-GKTVGDVVKQY 83
8*****************99*************.***99998877 PP
| |||||||
| 2 | Myb_DNA-binding | 44.8 | 2.9e-14 | 149 | 193 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE++l++ + k++G+g+W+ I+r + +Rt+ q+ s+ qky
Kaladp0036s0258.1.p 149 PWTEEEHKLFLLGLKKYGKGDWRNISRNFVITRTPTQVASHAQKY 193
8*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 7.661 | 36 | 91 | IPR017884 | SANT domain |
| SMART | SM00717 | 4.8E-7 | 37 | 89 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.27E-6 | 40 | 83 | No hit | No description |
| Pfam | PF00249 | 3.7E-6 | 40 | 85 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 2.44E-10 | 40 | 93 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-12 | 141 | 198 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.486 | 142 | 198 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.48E-17 | 143 | 197 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.4E-17 | 145 | 197 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-13 | 146 | 196 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.05E-11 | 149 | 194 | No hit | No description |
| Pfam | PF00249 | 1.0E-12 | 149 | 193 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 292 aa Download sequence Send to blast |
MKFEMEIRSP GYGYPENNQQ QNSRQNRKQQ HHQQAESLSK WSQEENKMFE NALAVYDRDS 60 PDRWQKVAAM IPGKTVGDVV KQYEDLEADV SSIEAGLVPV PGYACPAFTL DWVGGDGGGG 120 SKSYDGFSYG YGGGKRLRGV EQERKKGVPW TEEEHKLFLL GLKKYGKGDW RNISRNFVIT 180 RTPTQVASHA QKYFIRQLSG GKEKRRASIH DITTVNLSES QTPLSPDQSA RGQHSPEMPF 240 QWGGSQSSSG HGSSIDFNGV YGFSSNRSAQ GMLGSYMDAY QMQSATQYPQ V* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 1e-15 | 41 | 103 | 11 | 73 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
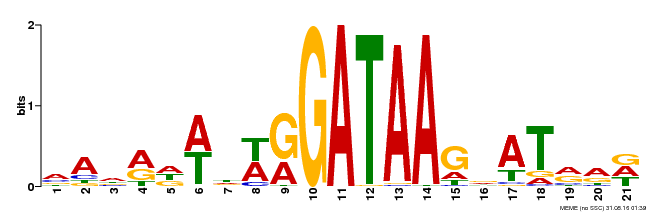
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00296 | DAP | Transfer from AT2G38090 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK362184 | 1e-42 | AK362184.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2003C15. | |||
| GenBank | AK363192 | 1e-42 | AK363192.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2013G01. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021278855.1 | 1e-109 | transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 5e-99 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | A0A061GWE3 | 1e-106 | A0A061GWE3_THECC; DIV1A protein | ||||
| STRING | EOY33801 | 1e-107 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58900.1 | 1e-78 | Homeodomain-like transcriptional regulator | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0036s0258.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



