 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0039s0692.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 421aa MW: 46450.3 Da PI: 8.3696 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 20.4 | 1.3e-06 | 256 | 277 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
C Cgk F+r nL+ H+r H
Kaladp0039s0692.1.p 256 FCRVCGKGFKRDANLRMHMRAH 277
6*******************99 PP
| |||||||
| 2 | zf-C2H2 | 11.5 | 0.00093 | 356 | 379 | 1 | 23 |
EEETTTT.EEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCg.ksFsrksnLkrHirtH 23
y C+ Cg k Fs s+L++H + +
Kaladp0039s0692.1.p 356 YACKRCGqKQFSVLSDLRTHEKHC 379
89******************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 8.74E-6 | 253 | 280 | No hit | No description |
| PROSITE profile | PS50157 | 12.404 | 255 | 282 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0037 | 255 | 277 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 257 | 277 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 4.2E-6 | 257 | 281 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 270 | 318 | 351 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 24 | 356 | 378 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 421 aa Download sequence Send to blast |
MAPTSSSCFL PNEPASSSQP MLGYGVMTSN GGHEDVESSN PVGLNHFSTS LLCSISILKE 60 RVRQAQSLVV SLFASPSITD QHYSQQVVEP ASMTMARISS LVQDIVAVST SMMLTCQQMS 120 LSNPTRQHFQ ASSDHHHPSP ISGHRHNQTL IRDGDGNNHV IIQAGQDVGH QAAHKQEQIL 180 GHRDMFGIGS SEQDMEWFGG SSCSFANATL SYDTATTSTT NSIQQESNNT EHHQLSHPDL 240 IIEMDAADLL AKYTHFCRVC GKGFKRDANL RMHMRAHGDE YKSIAALANP AKINNDDPNI 300 PATTPCTADK TARRSIRYSC PHHGCRWNRK HPKFQPLKSM ICAKNHYKRS HCPKMYACKR 360 CGQKQFSVLS DLRTHEKHCG ESKWRCSCGT TFSRKDKLLG HVGLFAGHVP VIVNHVLSHT 420 * |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00522 | DAP | Transfer from AT5G22890 | Download |
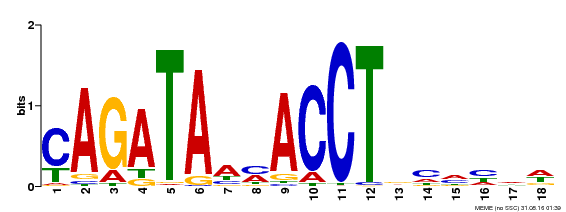
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009356942.1 | 1e-114 | PREDICTED: zinc finger protein STOP1 homolog | ||||
| TrEMBL | A0A498INF8 | 1e-111 | A0A498INF8_MALDO; Uncharacterized protein | ||||
| STRING | XP_009356942.1 | 1e-113 | (Pyrus x bretschneideri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22890.1 | 2e-94 | C2H2 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0039s0692.1.p |



