 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0048s0343.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 348aa MW: 39143.8 Da PI: 8.2341 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 100.9 | 9e-32 | 111 | 166 | 3 | 59 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRrevee 59
+vrY+eClkNhAas+G+++vDGCgEfmp g+eg+ +alkCaAC+CHRnFHRre e
Kaladp0048s0343.1.p 111 GVRYRECLKNHAASMGTNVVDGCGEFMPG-GKEGSLEALKCAACQCHRNFHRRELEG 166
79**************************9.999********************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 5.0E-20 | 106 | 165 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 4.3E-29 | 112 | 164 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 1.5E-29 | 113 | 163 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 25.731 | 114 | 163 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-30 | 234 | 305 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.21E-18 | 239 | 305 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 2.3E-28 | 244 | 301 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 348 aa Download sequence Send to blast |
MEQYQQQQQQ QHNQIKLSQH HTTTPSPPRN LPPPPPLAYP PSSIEQPPRT HTSLHDRDPE 60 VEDKKNGVLF LSAPSAGMYG GGGGELQSPE SLKLSPVGTR SKPVKSASPS GVRYRECLKN 120 HAASMGTNVV DGCGEFMPGG KEGSLEALKC AACQCHRNFH RRELEGHESG SSNFARSVIV 180 HPLHLPPPTQ IPLHHHHPHH HHHHQIVQPV AVAYGGGATE SSSEDLVQAL DEMPQREAEP 240 GSVRKRFRTK FTQEQKDKML GFAERLGWRL QRQDDEVVER FCSEVGVRRQ VLKVWMHNNK 300 NSFKKPPDED QQQQTHNHPP TTHPQIQLQP QHHHLLQLHQ QKQQETL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh7_A | 8e-27 | 245 | 302 | 18 | 75 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00258 | DAP | Transfer from AT2G02540 | Download |
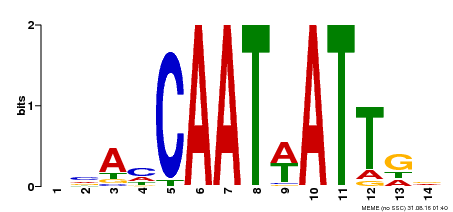
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022951673.1 | 9e-78 | zinc-finger homeodomain protein 2-like | ||||
| Swissprot | O64722 | 1e-64 | ZHD3_ARATH; Zinc-finger homeodomain protein 3 | ||||
| TrEMBL | A0A2N9GME9 | 2e-72 | A0A2N9GME9_FAGSY; Uncharacterized protein | ||||
| STRING | XP_010539899.1 | 6e-73 | (Tarenaya hassleriana) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02540.1 | 1e-56 | homeobox protein 21 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0048s0343.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



