 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0079s0185.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 541aa MW: 59993.7 Da PI: 7.1856 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 50.4 | 5.6e-16 | 277 | 336 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g ++ ++++lg ++ +e+Aa+a++ a++k++g
Kaladp0079s0185.1.p 277 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqsRKgRQVYLGGYDMEEKAARAYDLAALKYWG 336
78*******************988886678446*************************98 PP
| |||||||
| 2 | AP2 | 46.8 | 7.3e-15 | 379 | 430 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV+++++ grW A+I + +k +lg+f t eeAa+a++ a+ k++g
Kaladp0079s0185.1.p 379 SIYRGVTRHHQHGRWQARIGRVAG---NKDLYLGTFSTQEEAAEAYDIAAIKFRG 430
57****************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 9.0E-13 | 277 | 336 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.55E-22 | 277 | 345 | No hit | No description |
| SuperFamily | SSF54171 | 1.11E-16 | 277 | 346 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 4.6E-16 | 278 | 345 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.137 | 278 | 344 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.1E-29 | 278 | 350 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.6E-6 | 279 | 290 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.9E-17 | 379 | 439 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 1.82E-25 | 379 | 440 | No hit | No description |
| Pfam | PF00847 | 1.1E-9 | 379 | 430 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.7E-18 | 380 | 438 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 18.927 | 380 | 438 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.3E-34 | 380 | 444 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.6E-6 | 420 | 440 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 541 aa Download sequence Send to blast |
MKCSMSNENA AAAATTNINH TSWLGFSLAP RMTMELPLDS HHQPVPYSSA APTLPSTYFH 60 TLPNLDASRN CYGLENGDYS SSGFPAMPLN SDGSLCIMGA SMCTVSLSHA PKLEDFFGGG 120 TMGSRECEAK DRQAAMALSL DSTCYQQQTT DQETSTSLHF LHNLQDGMRP QQIEIQHHPL 180 YCLPSSLGDD DHYELKNCSN DDALGERVNH VGFGDLQALT LSMSPGSQSG NAFASQRVPH 240 PAVEPLWLDS KKRGVEKVNQ KPIVHRKSID TFGQRTSQYR GVTRHRWTGR YEAHLWDNSC 300 KKEGQSRKGR QVYLGGYDME EKAARAYDLA ALKYWGSSTH INFPVENYQQ ELEAMKNMSR 360 QEYVANLRRK SSGFSRGASI YRGVTRHHQH GRWQARIGRV AGNKDLYLGT FSTQEEAAEA 420 YDIAAIKFRG VSAVTNFDIT RYNVERIIES STLPSGDLAR RSKEPEPITA HAADQHLTNT 480 ESPNIASPGS QHDSMSFHQM PPQLTAHHKP QLQHYSAVIS ESDEVANYVS KAPSQVTSFK 540 * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CAC[AG]N[AT]TNCCNANG-3'. Required for the initiation and growth of ovules integumenta, and for the development of female gametophyte. Plays a critical role in the development of gynoecium marginal tissues (e.g. stigma, style and septa), and in the fusion of carpels and of medial ridges leading to ovule primordia. Also involved in organs initiation and development, including floral organs. Maintains the meristematic competence of cells and consequently sustains expression of cell cycle regulators during organogenesis, thus controlling the final size of each organ by controlling their cell number. Regulates INO autoinduction and expression pattern. As ANT promotes petal cell identity and mediates down-regulation of AG in flower whorl 2, it functions as a class A homeotic gene. {ECO:0000269|PubMed:10528263, ECO:0000269|PubMed:10639184, ECO:0000269|PubMed:10948255, ECO:0000269|PubMed:11041883, ECO:0000269|PubMed:12183381, ECO:0000269|PubMed:12271029, ECO:0000269|PubMed:12655002, ECO:0000269|PubMed:8742706, ECO:0000269|PubMed:8742707, ECO:0000269|PubMed:9001406, ECO:0000269|PubMed:9093862, ECO:0000269|PubMed:9118807}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
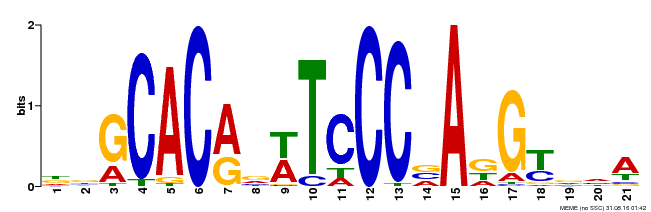
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021682654.1 | 0.0 | AP2-like ethylene-responsive transcription factor ANT | ||||
| Swissprot | Q38914 | 1e-144 | ANT_ARATH; AP2-like ethylene-responsive transcription factor ANT | ||||
| TrEMBL | A0A1U8B2W5 | 0.0 | A0A1U8B2W5_NELNU; AP2-like ethylene-responsive transcription factor ANT | ||||
| STRING | XP_010273592.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-139 | AP2 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0079s0185.1.p |



