 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0081s0001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 370aa MW: 42324 Da PI: 8.3475 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 18.9 | 4e-06 | 67 | 88 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C+ Cg F+ + +Lk+H+ +H
Kaladp0081s0001.1.p 67 TCQECGANFKKPAYLKQHMLSH 88
6*******************99 PP
| |||||||
| 2 | zf-C2H2 | 15.7 | 4.3e-05 | 94 | 118 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp C+ s++rk++L+rH+ H
Kaladp0081s0001.1.p 94 FMCPvdGCHSSYRRKDHLTRHLLQH 118
78*******************9887 PP
| |||||||
| 3 | zf-C2H2 | 16.7 | 2e-05 | 160 | 184 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ Cgk F+ s L++H +H
Kaladp0081s0001.1.p 160 HVCQedGCGKAFRFASKLQKHELSH 184
789999***************9998 PP
| |||||||
| 4 | zf-C2H2 | 15 | 7e-05 | 251 | 275 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C +Fs++snL++H + H
Kaladp0081s0001.1.p 251 KCNfeGCLHTFSNRSNLQQHVKAvH 275
699999**************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 74 | 23 | 46 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.064 | 66 | 88 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.757 | 66 | 93 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.4E-7 | 66 | 87 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 68 | 88 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.43E-10 | 80 | 120 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-12 | 88 | 120 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0072 | 94 | 118 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.12 | 94 | 123 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 96 | 118 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 43 | 123 | 146 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.556 | 123 | 151 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-7 | 157 | 184 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.02E-5 | 157 | 186 | No hit | No description |
| PROSITE profile | PS50157 | 10.45 | 160 | 189 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.023 | 160 | 184 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 162 | 184 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.2 | 192 | 217 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 194 | 217 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 11 | 220 | 241 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 9.0E-9 | 231 | 272 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 8.44E-8 | 231 | 288 | No hit | No description |
| SMART | SM00355 | 0.0024 | 250 | 275 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.159 | 250 | 280 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 252 | 275 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5 | 281 | 306 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 283 | 307 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 370 aa Download sequence Send to blast |
MEETQQQGEE SAKQPSFRDI RRYYCEYCGV CRSKKSLISM HIIKFHQKEM DENDGEDNIE 60 ETAKTNTCQE CGANFKKPAY LKQHMLSHSV ERPFMCPVDG CHSSYRRKDH LTRHLLQHEG 120 NVFKCSNEKC ARQFVSQANL TRHKCHEPET PFSGCASKQH VCQEDGCGKA FRFASKLQKH 180 ELSHVKLETM EAICLETGCL KYFSNEACLK AHMDACHQHI NCEVCGSKQL KKNLKRHLRT 240 HEPGHANEPV KCNFEGCLHT FSNRSNLQQH VKAVHLNIKR FACGYADCGM KFAYKHVRDN 300 HEKTSHHVYT QGDFLESLEH CRVTPCGGRK RKCPSIASVL VKRITPHPKD SDSTASSMYS 360 AWMNVTNSE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 3e-16 | 55 | 212 | 2 | 155 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 3e-16 | 55 | 212 | 2 | 155 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
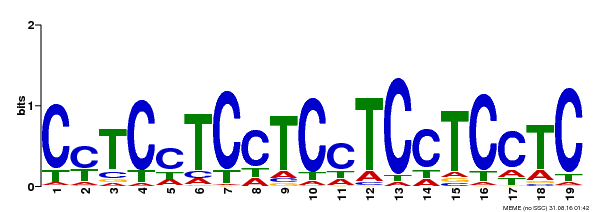
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010251838.1 | 1e-155 | PREDICTED: transcription factor IIIA | ||||
| Refseq | XP_010251839.1 | 1e-155 | PREDICTED: transcription factor IIIA | ||||
| Refseq | XP_010251840.1 | 1e-155 | PREDICTED: transcription factor IIIA | ||||
| Refseq | XP_010251841.1 | 1e-155 | PREDICTED: transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 1e-116 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A1U7ZTZ8 | 1e-154 | A0A1U7ZTZ8_NELNU; transcription factor IIIA | ||||
| STRING | XP_010251838.1 | 1e-155 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-118 | transcription factor IIIA | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0081s0001.1.p |



