 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0093s0085.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 459aa MW: 50008 Da PI: 7.7754 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 48.2 | 2e-15 | 286 | 331 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRd+iN+++ +L++l+P++ K +Ka++L ++++Y+k+Lq
Kaladp0093s0085.1.p 286 HNQSERKRRDKINQRMKTLQKLVPNS-----NKTDKASMLDEVIDYLKQLQ 331
9************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 9.95E-19 | 279 | 343 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.727 | 281 | 330 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.5E-13 | 286 | 331 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 9.2E-18 | 286 | 340 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.17E-16 | 286 | 334 | No hit | No description |
| SMART | SM00353 | 2.3E-17 | 287 | 336 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 459 aa Download sequence Send to blast |
MSQCVPSWDN NPHNDTYSHT LHPHLPPAPA KQLVHSRNLD SGIVPSSAPE VPAMLDYEVA 60 ELTWENGQLA LHGLGQPRTS GAWQDKHQPG SGTLESIVNQ ATCFFNPNDK SSFDHRAPTA 120 SAAMVMDALV PLCSNTNNNN CNCNNNNNNG NNPDEDSNAR GTVGHTHVGS CSAGMKRTRQ 180 GVRQVPVVGQ DWSARSEYSA NSSRQQQVTV DTCTERDQGT TAFTSTSVWS AENTSSGMPC 240 TRLTGDDHDS ICHSRSIMKN ERGSGENKKQ KASNNSAKRS RAADIHNQSE RKRRDKINQR 300 MKTLQKLVPN SNKTDKASML DEVIDYLKQL QAQVNMMNRM NMPQMAMMPM ALNQQAALQM 360 TMMAQMGMGG MLDMNTAAAM AAGRASMQPI LHPSAAAAAF MPMPPASWDA AAQAMMSDPM 420 SAFLACQQSQ PMTMDAYRMM ASLYQHMQQA QQAPNSKS* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 289 | 294 | ERKRRD |
| 2 | 290 | 295 | RKRRDK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
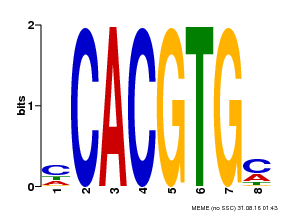
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011017451.1 | 1e-109 | PREDICTED: transcription factor UNE10-like | ||||
| TrEMBL | U5GLG2 | 1e-106 | U5GLG2_POPTR; BHLH family protein | ||||
| STRING | POPTR_0002s14430.1 | 1e-107 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 7e-62 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0093s0085.1.p |



