 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0093s0085.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 336aa MW: 36694.4 Da PI: 8.943 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 48 | 2.2e-15 | 163 | 208 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRd+iN+++ +L++l+P++ K +Ka++L ++++Y+k+Lq
Kaladp0093s0085.2.p 163 HNQSERKRRDKINQRMKTLQKLVPNS-----NKTDKASMLDEVIDYLKQLQ 208
9************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 6.02E-19 | 156 | 220 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.727 | 158 | 207 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.7E-18 | 163 | 217 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 6.1E-13 | 163 | 208 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.17E-15 | 163 | 211 | No hit | No description |
| SMART | SM00353 | 2.3E-17 | 164 | 213 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MVMDALVPLC SNTNNNNCNC NNNNNNGNNP DEDSNARGTV GHTHVGSCSA GMKRTRQGVR 60 QVPVVGQDWS ARSEYSANSS RQQQVTVDTC TERDQGTTAF TSTSVWSAEN TSSGMPCTRL 120 TGDDHDSICH SRSIMKNERG SGENKKQKAS NNSAKRSRAA DIHNQSERKR RDKINQRMKT 180 LQKLVPNSNK TDKASMLDEV IDYLKQLQAQ VNMMNRMNMP QMAMMPMALN QQAALQMTMM 240 AQMGMGGMLD MNTAAAMAAG RASMQPILHP SAAAAAFMPM PPASWDAAAQ AMMSDPMSAF 300 LACQQSQPMT MDAYRMMASL YQHMQQAQQA PNSKS* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 166 | 171 | ERKRRD |
| 2 | 167 | 172 | RKRRDK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
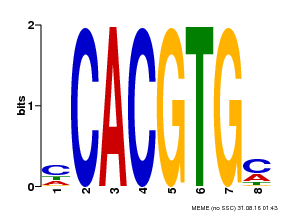
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011017451.1 | 1e-82 | PREDICTED: transcription factor UNE10-like | ||||
| TrEMBL | A0A2G9GTZ3 | 6e-81 | A0A2G9GTZ3_9LAMI; Transcriptional repressors of the hairy/E(Spl) family (Contains HLH) | ||||
| STRING | POPTR_0002s14430.1 | 1e-80 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 1e-37 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0093s0085.2.p |



