 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0095s0014.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 281aa MW: 31441.7 Da PI: 6.5133 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 78.1 | 6.2e-25 | 60 | 110 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
k+i+n++ rqvtfskRr g++KKAeEL vLCd +v +++fsstgk +eys+
Kaladp0095s0014.1.p 60 KKIDNTTARQVTFSKRRRGLFKKAEELAVLCDSDVGLVVFSSTGKCFEYSN 110
68***********************************************95 PP
| |||||||
| 2 | K-box | 41.7 | 5.1e-15 | 146 | 225 | 20 | 99 |
K-box 20 elakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkkle 99
+++kL +e+ + + +R++ Ge+Le + +++LqqLe+ Le +l++i ++K ++++i el +k +l een+ Lr++++
Kaladp0095s0014.1.p 146 NYTKLGQEVAEKSSLLRRYRGENLEGMGIEDLQQLEKCLEAGLSRIIETKGGKIMREICELENKGMRLMEENEVLRQQVA 225
6788888888888999*************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF55455 | 7.33E-28 | 52 | 122 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 28.132 | 52 | 112 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 2.0E-35 | 52 | 111 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 9.01E-36 | 53 | 128 | No hit | No description |
| PRINTS | PR00404 | 4.5E-25 | 54 | 74 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 54 | 108 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 2.4E-24 | 61 | 108 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.5E-25 | 74 | 89 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.5E-25 | 89 | 110 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 11.876 | 140 | 230 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 1.0E-14 | 146 | 224 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009266 | Biological Process | response to temperature stimulus | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010076 | Biological Process | maintenance of floral meristem identity | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048438 | Biological Process | floral whorl development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000900 | Molecular Function | translation repressor activity, nucleic acid binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 281 aa Download sequence Send to blast |
MFLHFDWLAS LSLSLSLFSP HPENTVGSNC NGGHGSSTPA ARLEREKRVS EMAREKIQIK 60 KIDNTTARQV TFSKRRRGLF KKAEELAVLC DSDVGLVVFS STGKCFEYSN CGMQQILERH 120 HLQSKNLEKL QQPAPSLELQ LVEDVNYTKL GQEVAEKSSL LRRYRGENLE GMGIEDLQQL 180 EKCLEAGLSR IIETKGGKIM REICELENKG MRLMEENEVL RQQVARIPNV YIPHRANSEN 240 RMCEDGQSSE SVTNLCISGG ASQDYESSDI SLRLGLPYSG * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 3e-17 | 52 | 120 | 1 | 69 | MEF2C |
| 5f28_B | 3e-17 | 52 | 120 | 1 | 69 | MEF2C |
| 5f28_C | 3e-17 | 52 | 120 | 1 | 69 | MEF2C |
| 5f28_D | 3e-17 | 52 | 120 | 1 | 69 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that inhibit floral transition in the autonomous flowering pathway, independent of photoperiod and temperature. Acts in a dosage-dependent manner. Together with AGL24 and AP1, controls the identity of the floral meristem and regulates expression of class B, C and E genes. Promotes EFM expression to suppress flowering (PubMed:25132385). {ECO:0000269|PubMed:16679456, ECO:0000269|PubMed:18694458, ECO:0000269|PubMed:19656343, ECO:0000269|PubMed:25132385}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00272 | DAP | Transfer from AT2G22540 | Download |
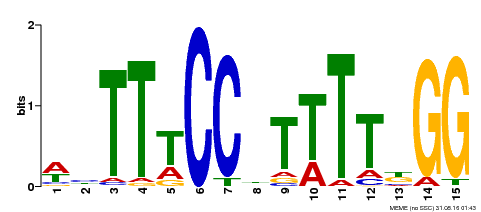
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the floral homeotic genes AP1 and SEP3 in emerging floral meristems. Up-regulated by HUA2. {ECO:0000269|PubMed:15659097, ECO:0000269|PubMed:17428825, ECO:0000269|PubMed:18694458}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021662492.1 | 1e-107 | MADS-box protein SVP | ||||
| Swissprot | Q9FVC1 | 2e-95 | SVP_ARATH; MADS-box protein SVP | ||||
| TrEMBL | A0A2I4QQS2 | 1e-105 | A0A2I4QQS2_HEVBR; HbMADS-box protein | ||||
| STRING | VIT_00s0313g00070.t01 | 1e-105 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22540.1 | 1e-97 | MIKC_MADS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0095s0014.1.p |



