 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kaladp0100s0015.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 953aa MW: 105322 Da PI: 5.7356 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 80.4 | 1.9e-25 | 147 | 248 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFk 83
f+k+lt sd++++g +++p++ ae+ ++++ + ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv+a++L++gD+v+F
Kaladp0100s0015.1.p 147 FCKTLTASDTSTHGGFSVPRRAAEKLfptldytMQPP-T-QELVVRDLHENTWTFRHIYRGQPKRHLLTTGWSLFVSAKRLRQGDSVLFI 234
99*********************999*****954444.4.38************************************************ PP
E-SSSEE..EEEEE-S CS
B3 84 ldgrsefelvvkvfrk 99
++++ +l+v+++r
Kaladp0100s0015.1.p 235 --RDEKSQLMVGIRRV 248
..4577778****996 PP
| |||||||
| 2 | Auxin_resp | 106.1 | 4.1e-35 | 273 | 356 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aa aa+++s+F+++YnPr+++s+Fv++++k++ka+ +++s+GmRf m+fete+s +rr++Gt+v v+dldp+rWp+SkWr+++
Kaladp0100s0015.1.p 273 AAYAAANRSPFTIFYNPRQCPSDFVIPMAKFRKAVFgTQISMGMRFGMMFETEESGKRRYMGTIVNVTDLDPLRWPGSKWRNIQ 356
6889******************************988********************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 2.62E-46 | 135 | 275 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.2E-41 | 140 | 261 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 9.63E-24 | 146 | 247 | No hit | No description |
| Pfam | PF02362 | 3.8E-23 | 147 | 248 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.14 | 147 | 249 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 3.4E-26 | 147 | 249 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.7E-30 | 273 | 356 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 2.6E-9 | 829 | 917 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 25.747 | 831 | 915 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 1.57E-8 | 835 | 910 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 953 aa Download sequence Send to blast |
MGSVVEEKIR PGGLPDGAST NLLEEMKMLK ELQDQSGAAK IMNSELWHAC AGPLVSLPQI 60 GGLVYYFPQG HSEQVAVSTK RSATSQIPNY PNLPPQLLCQ VHNVTLHADK ETDEIYAQMS 120 LQPVNTEKDI IPVPEFGRPS KHPSEFFCKT LTASDTSTHG GFSVPRRAAE KLFPTLDYTM 180 QPPTQELVVR DLHENTWTFR HIYRGQPKRH LLTTGWSLFV SAKRLRQGDS VLFIRDEKSQ 240 LMVGIRRVNR QQTTLPSSVL SADSMHIGVL AAAAYAAANR SPFTIFYNPR QCPSDFVIPM 300 AKFRKAVFGT QISMGMRFGM MFETEESGKR RYMGTIVNVT DLDPLRWPGS KWRNIQVEWD 360 EAGCSDKPNR VSSWDIETPE SLFIFPSLTS GYKRPLTPGF LGAETDWGSL IKRPFVRMPD 420 NGFGNLHPMA SPCSEQLMKM LMKSQMVNHP SNFSAIPVDS SYNRAPVPDG RAMEAVTKQN 480 PDQPMFSRNT LAQIQSSTQP CPAQSAPANS KVLPQVTHQE KTLQVVKSEI ASSEPVAQDH 540 GNQLKSGPHT VENFAPSSMN PPTSVTQPAS LNQQTTQRIL PNPSQAQFQQ VNPWQPHQPM 600 DSSIFQSQPM EAPQTDLSSI NSLLPYLETD EWMLNTPSSY QQLPGVFRSP ASLPIYGYQD 660 SSVVFPEVVN PVMSSIGQDV SEHQLNNVQS LSESDRLIPL PLQGQVNVSS SGFKDLSSDN 720 NPSNSRGVYG CLNYDGRNTS TAVVDHSVPS TFLDGFCKMD DPGFQNAPDC LVSSSQDLQS 780 QITSASLADS HAFSRQDFPD NSGGTSSSNV EFDDSSLLQT NTWQQVAPPM RTYTKVQKTG 840 SVGRSIDVTS FRNYEELRSA IERMFGLEGL LTDPRGSAWK LVYVDYEKDV LLVGDDPWEE 900 FVGCVRCIRI LSPTEVQQMS EEGMKLLNNT AAMQGLRRAL PEGGGSAWDA LA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 3 | 381 | 6 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
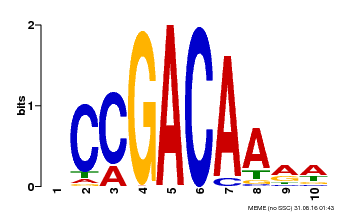
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021285434.1 | 0.0 | auxin response factor 5 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | A0A061FD42 | 0.0 | A0A061FD42_THECC; Auxin response factor | ||||
| STRING | EOY14976 | 0.0 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kaladp0100s0015.1.p |



