 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0023s0004.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 683aa MW: 77381.8 Da PI: 7.6292 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 37.9 | 4.8e-12 | 160 | 215 | 2 | 60 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkr 60
W+++ev aL+++r++ e++ We+vs+k+++ gf+rs+k+Ckek+e+ +k
Kalax.0023s0004.1.p 160 WSNEEVVALLRIRSSSETSWFPP---DFTWEHVSRKLAAIGFKRSAKTCKEKFEEESKT 215
**************999887655...699*************************98874 PP
| |||||||
| 2 | trihelix | 89.9 | 2.8e-28 | 561 | 656 | 1 | 86 |
trihelix 1 rWtkqevlaLiearr.........emeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkr.tsessstcp 80
rW++++vlaLi++r + e++ +++++++plWe++s+km++ g++rs+k+Ckekwen+nk+++k+k+++ ++ +s++s+tcp
Kalax.0023s0004.1.p 561 RWPRDQVLALINLRCslltnkngdNEERDEGNKQQAAPLWERISRKMSQLGYQRSAKRCKEKWENINKYFRKTKDNNVNKkRSNDSRTCP 650
8*************966666554333333334569*****************************************9888579******* PP
trihelix 81 yfdqle 86
yf+ql
Kalax.0023s0004.1.p 651 YFHQLS 656
****96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 3.7 | 156 | 215 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 3.6E-8 | 159 | 239 | No hit | No description |
| PROSITE profile | PS50090 | 5.982 | 160 | 213 | IPR017877 | Myb-like domain |
| PROSITE profile | PS50090 | 6.179 | 554 | 627 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 2.5 | 558 | 629 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 9.4E-19 | 560 | 658 | No hit | No description |
| CDD | cd12203 | 1.81E-22 | 561 | 634 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 683 aa Download sequence Send to blast |
MFDGMHNQDQ LFHQFISFSS PSSSRPPPLP SSLHHQQISS PFPASFPVAS PPPIIPPPPP 60 PNQSIQFSDL FHRPTNLPSF HHLNPAPSSH NLISSKQYTS DQHTNSNSIS DVLEAQVHPK 120 DEVGLIRSLF PRAAASSSSL LVERERPSSS PPTLHQHEFW SNEEVVALLR IRSSSETSWF 180 PPDFTWEHVS RKLAAIGFKR SAKTCKEKFE EESKTCFDTF NNRVISDRIS YRIFSELENI 240 CGRDDNNAEK SAIQLQANDR EEDPIININN HQDMQAEEEE GIACENKANT RCNVFEEDAN 300 CVSPAIIIQQ YSQREQLECR EKVMKAKAKK KKKKIRRNKQ EHKFQIMKGL CEGLVNKMMT 360 QQEELHNKIL EDLLKRDEEN KAREEAWKRM EVERLNTELE IRAQEQAIAG DRQAAIIDLL 420 GTTTTSATSH ESRRGIIDAN IVAHLLKPIP HMPTKVITQS HDQNPPPPDQ PPCPNLQTLI 480 AHEALEPPQT PTSTIEPHIS NPPACLPASY STTPADSSGG RSASAQLICY PKPSTKKAII 540 QQPNNNSRAG ILSEEDQHGV RWPRDQVLAL INLRCSLLTN KNGDNEERDE GNKQQAAPLW 600 ERISRKMSQL GYQRSAKRCK EKWENINKYF RKTKDNNVNK KRSNDSRTCP YFHQLSQLYE 660 QGTLTAPSHH QTSLLENGQN KM* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 328 | 336 | KKKKKKIRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
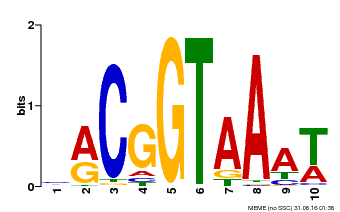
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 5e-65 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0023s0004.1.p |



