 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0065s0107.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 655aa MW: 71740.9 Da PI: 6.9617 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 62.3 | 8.1e-20 | 515 | 612 | 2 | 96 |
EEE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEE.TTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-S CS
B3 2 fkvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltled.esgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFkldg 86
+kvl++sdv+++gr+vlpkk ae+h ++ + ++ + +ed ++++W++++++ ++ks++y+l+ ++ +Fv++ngL+egDf+v++ +
Kalax.0065s0107.1.p 515 QKVLKQSDVGSLGRIVLPKKEAETHlpQLTARDGINIAMEDiGTSNVWNMRYRFwpNNKSRMYLLE-NTGDFVRSNGLQEGDFIVIY--S 601
79*************************999999********7778*******99777777777777.********************..6 PP
S.SEE..EEEE CS
B3 87 r.sefelvvkv 96
+ ++++++++
Kalax.0065s0107.1.p 602 DiKCGKYMIRG 612
77999888765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 2.9E-27 | 508 | 618 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10015 | 7.88E-34 | 512 | 613 | No hit | No description |
| SuperFamily | SSF101936 | 6.47E-22 | 513 | 607 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 10.306 | 514 | 616 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 6.5E-19 | 514 | 616 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 4.5E-16 | 515 | 611 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 655 aa Download sequence Send to blast |
MLDLSCFGGG GYDSAAAAME EADHQDLMDS AAAIFCDDLI PPLPDFQCMS SSSSSSSSVK 60 PVAYQSTASS SVSSTSSSWA VLRSESEEHN AAAGGYTSHH DHSHGHHPQQ VVREQSNGLD 120 EDGDIGCMDV METLSYIDLL DESGQVWGYK DSPVFYSSPV EAFQAENELQ RPETMAETHT 180 EGLDKSEDLA VVFFEWLKSN KESISAEDLR SIKLKKSTIE SAAKRLGNGK EGMKRLLQLI 240 LEWVQHHQLQ RKPSRAEGEA AAAAQPQFPN HPYPDTNNFQ PDLSHTSPWS VGSSAGLDPM 300 MAVGVPAPFG HLSNMADQPW PPLCMNYNNS ANAGQNLTPA AHPFGAAAFN QYPCQYYPAV 360 GNVSYVNGAV SEEVFVRTGS AATKEARKKR MARQRRFLPH HRHHHHHSSS GTGNAHGQHH 420 GSNGLSNQMR MLTGAGQLGR VNMDESCANI SHPPSGGYWA YLRNASPPQP PHFPVVPNSG 480 NSIQGNEESP SYQRPIALDR RQGSKPEKNL RFLLQKVLKQ SDVGSLGRIV LPKKEAETHL 540 PQLTARDGIN IAMEDIGTSN VWNMRYRFWP NNKSRMYLLE NTGDFVRSNG LQEGDFIVIY 600 SDIKCGKYMI RGVKVRQQGL KPDIRRSGKA LKKNTWPTSD GSPFAPPSPK TPLA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9b_A | 1e-37 | 510 | 618 | 1 | 109 | B3 domain-containing transcription factor FUS3 |
| 6j9b_D | 1e-37 | 510 | 618 | 1 | 109 | B3 domain-containing transcription factor FUS3 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
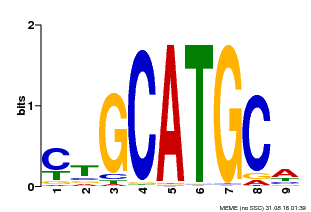
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021907276.1 | 1e-144 | B3 domain-containing transcription factor ABI3 | ||||
| TrEMBL | A0A1U7ZE45 | 1e-138 | A0A1U7ZE45_NELNU; B3 domain-containing transcription factor ABI3 | ||||
| STRING | evm.model.supercontig_8.187 | 1e-143 | (Carica papaya) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 1e-80 | B3 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0065s0107.1.p |



