 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0079s0014.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 791aa MW: 90929.6 Da PI: 8.2158 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 88 | 1.4e-27 | 58 | 145 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHela 90
+fY+eYAk++GF++++++s++sk+++e+++++f Cs++g + e + + ++r+ + ++t+Cka+++vkk+ gkw+v+++++eHnHel
Kalax.0079s0014.3.p 58 SFYQEYAKSMGFTTSIKNSRRSKKSKEFIDAKFACSRYGISPEPDCT---SSRRPTLKKTNCKASMHVKKRPGGKWVVYSFVKEHNHELL 144
5*****************************************99998...7788899*******************************97 PP
FAR1 91 p 91
p
Kalax.0079s0014.3.p 145 P 145
5 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 3.6E-25 | 58 | 145 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 2.6E-22 | 265 | 357 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 10.324 | 546 | 581 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 8.7E-7 | 553 | 578 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 1.3E-6 | 556 | 583 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 791 aa Download sequence Send to blast |
MVDHVQENHT YDGLNTNEAA ALLHKYGGAV DILEGDALQI EGEPHGGIEF ECHEAAYSFY 60 QEYAKSMGFT TSIKNSRRSK KSKEFIDAKF ACSRYGISPE PDCTSSRRPT LKKTNCKASM 120 HVKKRPGGKW VVYSFVKEHN HELLPALAYH FRIHKNVKVA EKNNIDILHA VSERTRKMYV 180 EMSRHSSEFW SFEFLKTDTV SQFDRGLHLA LDVGDAQFIL EYFKHVKKDS PGFLYAIDLN 240 EEHRLRNLFW VDAKSRSDYS SFSDVISLES CYIKNNNKMP FVPFVGVNNH FQPILLGCAL 300 VADDSKSTFV WLMKTWLRAM GGLAPKVIIT DHDQAMKMAV EEVFSTSCHC FSLHNILEKA 360 SEILIHVIQC HPKFLQKFNK CIFKSWTEED FDMRWWKMIG RFELQEYEWI RLLYDHRKKW 420 VPTYMKGTFL GGMCPIQRSE SMSAFLDRYI HKKITLKEFV RQYGTILHNI FEDEALADFD 480 TWHKQPALKS PSPWEKQVSA LYTHAIFKKF QVEVLGVVGC HIEKVGDDGT TVTFKVQDCE 540 KNERFYVAWI EAKSEVYCSC RSFEYRGFLC RHAMIVLQIC GFSSIPSQYI LTRWTKDAKS 600 MQSSSIGAIR VEDRIQRYNY LCKRAIELCE EGSLSQDNYS VAIRFLVEAL KNCVNENDSN 660 KNVGESNNAS ILKEHHGSLA FKRNKKRIPN KKRRALSETE FSADTSSNLH QMVPLNLMEP 720 PHDAYYINQQ SLQGLLTNIA SGHDIFFGPQ QSTRSINNLD FRVQANFTYG VQENTGSSPS 780 HVHGSSSGRA * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
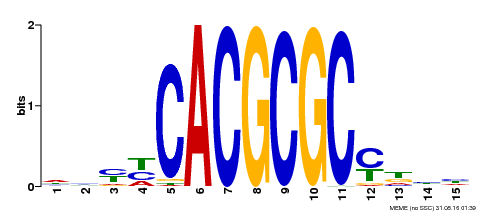
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010648203.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X2 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A2P6S0S3 | 0.0 | A0A2P6S0S3_ROSCH; Putative transcription factor FAR family | ||||
| STRING | VIT_03s0017g01810.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0079s0014.3.p |



