 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0146s0055.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 548aa MW: 60207.1 Da PI: 8.743 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 58.2 | 1.5e-18 | 256 | 305 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lre++P++ ++K +Ka+ L +A+eYI+ Lq
Kalax.0146s0055.3.p 256 RSKHSATEQRRRSKINDRFQMLREIIPHS----DQKRDKASFLLEAIEYIQFLQ 305
889*************************9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 4.6E-23 | 252 | 321 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.27E-18 | 254 | 313 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.072 | 254 | 304 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.08E-15 | 256 | 308 | No hit | No description |
| Pfam | PF00010 | 5.5E-16 | 256 | 305 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.9E-13 | 260 | 310 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 548 aa Download sequence Send to blast |
MELPRSFGPQ GTKPTHDFLS LYTTTSTTSH SQDPLLSPGS FLKTQNFLQP LEQVEMPGVN 60 DPAAISVNDK IQLPAASHLP PTSVEHILPG GIGTYTISHI SCINQVSKPE VSLFTAAQAN 120 SNERNDDNSN SSSQTGSGFT LWEESAVNMG KTGKENVREG RSFVREPASK LGKWAAERPS 180 QSSLNQRGSF APASSSQVAA HKNQSLMNLI VSTTNTREDE NREMDFVIKK EGSPSERESK 240 MRAEGKNDER KANTPRSKHS ATEQRRRSKI NDRFQMLREI IPHSDQKRDK ASFLLEAIEY 300 IQFLQEKVYR YEGSFPGWNH ESTKRIPLRN NHKPAKGFID HGGPGTTSPF STKFSDKNIG 360 APSIVSRVVQ KQVESDNNPI AFQMNDQHLV RADMIVASPI PLQPKVFVEN GGTTTIFPPK 420 LVSDTETATS QHTQREDVRR CEIECATASN DQELAIEGGT INISSTYSQG FLSTLTQALQ 480 SLGVDLSQSR ISVQIDLGKQ NNIKLGSATK MTKVDELNST SHVKANSTSP HISEYSGQAV 540 KRLKTGN* |
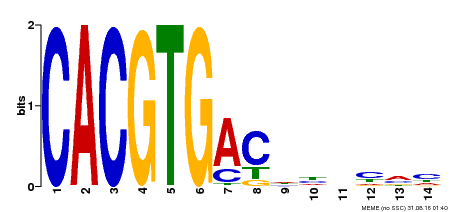
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019082147.1 | 1e-162 | PREDICTED: transcription factor BIM1 | ||||
| TrEMBL | D7SIP8 | 1e-158 | D7SIP8_VITVI; Uncharacterized protein | ||||
| STRING | VIT_17s0000g04790.t01 | 1e-159 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 2e-75 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0146s0055.3.p |



