 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0146s0062.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 672aa MW: 76175.5 Da PI: 6.0956 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 72.8 | 4.8e-23 | 202 | 255 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r+ Wt +LH++Fv+av+ G ek Pk+il+ m+v+ Lt+e+v+SHLQkYRl
Kalax.0146s0062.1.p 202 KARVIWTVDLHQKFVNAVNLV-GLEKIGPKKILDSMNVPWLTRENVASHLQKYRL 255
68*****************99.********************************8 PP
| |||||||
| 2 | Response_reg | 74.5 | 4.1e-25 | 22 | 130 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedale 88
vl+vdD+ + +++l+++l+k y ev+++ ++eal +l+e++ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + +
Kalax.0146s0062.1.p 22 VLVVDDDLTWLKILEKMLKKCFY-EVTTCCLAREALSILRERKdgFDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSIDGETSRVMK 109
89*********************.***************999999**********************6644.8***************** PP
HHHTTESEEEESS--HHHHHH CS
Response_reg 89 alkaGakdflsKpfdpeelvk 109
++ Ga d+l Kp+ ++el++
Kalax.0146s0062.1.p 110 GVQHGACDYLLKPIRMKELRN 130
******************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 3.0E-136 | 1 | 629 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 1.71E-34 | 19 | 147 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 3.1E-42 | 19 | 143 | No hit | No description |
| SMART | SM00448 | 5.3E-30 | 20 | 132 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 41.477 | 21 | 136 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.3E-22 | 22 | 131 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.65E-28 | 23 | 136 | No hit | No description |
| SuperFamily | SSF46689 | 5.38E-16 | 200 | 259 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-25 | 201 | 260 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.9E-21 | 202 | 255 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.8E-6 | 207 | 254 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 672 aa Download sequence Send to blast |
MMMEKSAFAS PQTDAFPAGL RVLVVDDDLT WLKILEKMLK KCFYEVTTCC LAREALSILR 60 ERKDGFDIVI SDVNMPDMDG FKLLEHVGLE MDLPVIMMSI DGETSRVMKG VQHGACDYLL 120 KPIRMKELRN IWQHVFRKRV HEVRDVENHE GFEAIQMVRI GCEQSDLEYY LSGGHDLSAT 180 RKRKDAVHKH NDRELGDAFA KKARVIWTVD LHQKFVNAVN LVGLEKIGPK KILDSMNVPW 240 LTRENVASHL QKYRLYLSRL HKYNGDEALE MANHLDLDFC RVKHSDELTK DSEEISFSTE 300 EPITEKHINS SSGASSSNSY YWGDHKIQDT KFRNFTLPDL KVEPIFGLNA DDPESQSMKY 360 SWPDASHRLE TVESDAQFAN SDPNITSKYS RSGEIRDIHY ELVNKQKQWL DDGFNSWSLP 420 NRQYHPQPNQ RSFLLSANTG SSHWEKEITS LSVEPDHLKP SLSAHVVKIL SEGETSVENN 480 PPSVEQGFHH DHVKGKAPLE HKFDTGFKAA HTSSLQTPVS HLLASPSVYL GTSNRETHTS 540 SSVKNKRPQS EQVTNSNRDT TSLETGSPML RAPTVGSFIA SQRFEYAESE LALVAFGKDY 600 KVNGDHGEYT SSNSYDFLPI TEPDIQANVP LHLYDAMRFD FCYEAAGFPM IDQSLSIVDT 660 AHFTSSYYPV S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 4e-16 | 201 | 260 | 4 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
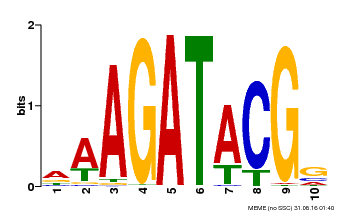
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028061797.1 | 1e-156 | two-component response regulator ORR26-like isoform X2 | ||||
| TrEMBL | A0A2R6RED2 | 1e-153 | A0A2R6RED2_ACTCH; Two-component response regulator like | ||||
| STRING | XP_002515016.1 | 1e-152 | (Ricinus communis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-115 | response regulator 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0146s0062.1.p |



