 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0171s0008.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 263aa MW: 28891.7 Da PI: 9.2441 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 127.4 | 6.8e-40 | 64 | 163 | 1 | 100 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleql 90
+CaaCk+lrr+Ca++C++ pyf ++p+kfa vhk+FGasnv+k+l ++pe +r d+++slv+eA++r+rdPvyG++g i++lqqq+++l
Kalax.0171s0008.1.p 64 PCAACKLLRRRCAQECPFFPYFSPHEPQKFASVHKVFGASNVSKMLMEVPEAHRADTANSLVFEANVRLRDPVYGCMGAISALQQQVQSL 153
7***************************************************************************************** PP
DUF260 91 kaelallkee 100
+ae++++++e
Kalax.0171s0008.1.p 154 QAEINAIRAE 163
*****99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 24.472 | 63 | 164 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 5.0E-40 | 64 | 161 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 263 aa Download sequence Send to blast |
MSRERDKSSQ FSDELEKKIK RSASLAATET ITSEDNSASD QMIVGRRQML GSPGRRTRLN 60 TITPCAACKL LRRRCAQECP FFPYFSPHEP QKFASVHKVF GASNVSKMLM EVPEAHRADT 120 ANSLVFEANV RLRDPVYGCM GAISALQQQV QSLQAEINAI RAEILNYKLR GEANGIITTN 180 SHSNCYMTSN FIPPSPPWAN ITVASLTPRP PSSSVVAATS ASHLLTTTTS SSLSVYTNQS 240 SIRNVMMGYS NSIVNENFRY LD* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 4e-44 | 64 | 178 | 11 | 124 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 4e-44 | 64 | 178 | 11 | 124 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00283 | DAP | Transfer from AT2G30340 | Download |
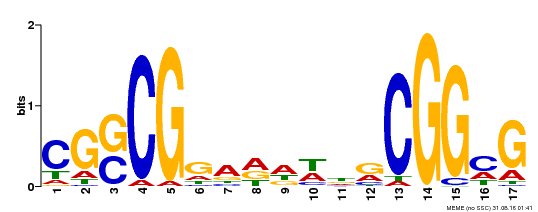
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023925198.1 | 2e-84 | LOB domain-containing protein 15 | ||||
| Swissprot | Q9AT61 | 2e-67 | LBD13_ARATH; LOB domain-containing protein 13 | ||||
| TrEMBL | A0A2P5AHJ8 | 5e-84 | A0A2P5AHJ8_TREOI; Wilm's tumor protein | ||||
| STRING | XP_010067591.1 | 4e-82 | (Eucalyptus grandis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40470.1 | 5e-68 | LOB domain-containing protein 15 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0171s0008.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



