 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0187s0047.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 422aa MW: 45722.7 Da PI: 9.7987 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51 | 3.4e-16 | 55 | 99 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a k++G W+ I +++g ++t+ q++s+ qk+
Kalax.0187s0047.1.p 55 REKWTEEEHQKFIEALKLYGRA-WRQIEEYIG-TKTAVQIRSHAQKF 99
789*****************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.42E-15 | 49 | 105 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.447 | 50 | 104 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-9 | 52 | 106 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-16 | 53 | 102 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.9E-13 | 54 | 102 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.7E-14 | 55 | 98 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.91E-10 | 57 | 100 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 422 aa Download sequence Send to blast |
MAPLQIPTAG SKSPVTSIAS GNNSQNILNA NCETDLDGSG VALKVRKPYT ISKQREKWTE 60 EEHQKFIEAL KLYGRAWRQI EEYIGTKTAV QIRSHAQKFF SKVTKESASS AEAPTEAIDI 120 PPPRPKRKPL HPYPRKFVEA PNKTRQGPSL KENGSSTISK NLERGALSPT SVLSPEVAHM 180 STSHAKAQGS GCVSATSSGS RSSSVLEKSN PNKNNSTPIK EKDLLRTLNL SGPSSHDSTS 240 VQTRKEFKYI ELTKDEDVYS EAETKFFGGV SPLKTIKLFG QTVVVTKPPD LSLDMKNVVM 300 PTATRGPLMC CVSHMDSLVG SQPYDSSPDG QLVTGGILMG ERSREKGGSK GFSVNGSISD 360 AISKAYNAER NQDTVDSKIR SSEGPILSPS EGRKGFVPYK RCVSEREAVL QMNKADCRVC 420 L* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
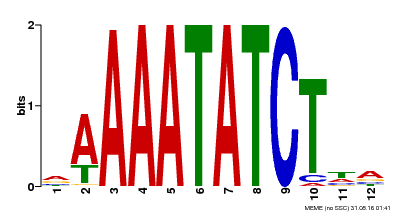
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G37260.1 | 2e-33 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0187s0047.1.p |



