 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0267s0057.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | M-type_MADS | ||||||||
| Protein Properties | Length: 276aa MW: 30733.2 Da PI: 9.8722 | ||||||||
| Description | M-type_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 49.7 | 4.6e-16 | 10 | 51 | 2 | 43 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TT CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCdaevaviifsst 43
+i n+s r+ t+ kR+ g++KK +ELS+LC+++++ +i+s+
Kalax.0267s0057.1.p 10 FITNDSARKATYKKRKRGLIKKVNELSTLCGVDACAVIYSPY 51
89**************************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 16.442 | 1 | 49 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 2.0E-22 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 1.7E-23 | 1 | 86 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00266 | 1.58E-32 | 2 | 86 | No hit | No description |
| PRINTS | PR00404 | 2.0E-7 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.3E-17 | 10 | 51 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.0E-7 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0043078 | Cellular Component | polar nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 276 aa Download sequence Send to blast |
MTRQKVKLAF ITNDSARKAT YKKRKRGLIK KVNELSTLCG VDACAVIYSP YETQPEVWPQ 60 SAVGVQRVLA QFRRMPEMEQ SKKMVNQETF LRQRIVKTKE QLKKQLKDNR EKEVTEVMFQ 120 CLTGKGMANL NMMDLNDLAW LIDQYMREIN NRCEKLKKSE VATGLQASGA GGADLVEEDT 180 KPLILDHVSR PSMMNSGVHH AAAGDGRQGS AWFPGGGFVS NNNGASSSSS SNKNNHLQLQ 240 VLPGIMADTD MILAPPQVNM GGMWPHQNPN FYNNM* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Controls central cell differentiation during female gametophyte development. Required for the expression of DEMETER and DD46, but not for the expression of FIS2 (PubMed:16798889). Probable transcription factor that may function in the maintenance of the proper function of the central cell in pollen tube attraction (Probable). {ECO:0000269|PubMed:16798889, ECO:0000305|PubMed:26462908}. | |||||
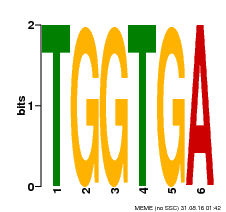
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00553 | DAP | Transfer from AT5G48670 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016464483.1 | 7e-82 | PREDICTED: agamous-like MADS-box protein AGL80 | ||||
| Swissprot | Q9FJK3 | 1e-64 | AGL80_ARATH; Agamous-like MADS-box protein AGL80 | ||||
| TrEMBL | A0A1S3ZJU3 | 2e-80 | A0A1S3ZJU3_TOBAC; agamous-like MADS-box protein AGL80 | ||||
| STRING | POPTR_0005s00420.1 | 3e-81 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G48670.1 | 1e-66 | AGAMOUS-like 80 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0267s0057.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



