 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0277s0041.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 318aa MW: 34301.6 Da PI: 9.4007 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 39.7 | 1.2e-12 | 60 | 104 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r++WT+ E++++++a +++ + Wk+I + +g +t q++s+ qky
Kalax.0277s0041.1.p 60 RQSWTEPEHDKFLEALQLFDRD-WKKIEAFIG-SKTVIQIRSHAQKY 104
899*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.5E-16 | 54 | 110 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.023 | 55 | 109 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 5.9E-18 | 58 | 107 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-6 | 58 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.9E-10 | 59 | 107 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.6E-10 | 60 | 104 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.31E-7 | 62 | 105 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 318 aa Download sequence Send to blast |
MVSNSSNPPD AFYLDPTGAA LPGLSPFAPT GDGGAIGVAS AAGEDGDKKV RKPYTITKSR 60 QSWTEPEHDK FLEALQLFDR DWKKIEAFIG SKTVIQIRSH AQKYFLKVQK SGGNEHLPPP 120 RPKRKAAHPY PHKASKSALP ELRGALLSSA SASSLPHPGY VLKHDVHSIP SDPSSRAAVA 180 SSWTDSSAQA SAIPNASQGD LKSTAEAMAI NSNNSAESNL RVQSARRTPV LANKSQAIRV 240 LPDFAQVYGF IGSVFDPNVS GHLQNLKQMD PIDVETVLLL MRNLSINLTS PAFEDHRKIL 300 STYEFDPKEP NYDVIGI* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00625 | PBM | Transfer from PK02532.1 | Download |
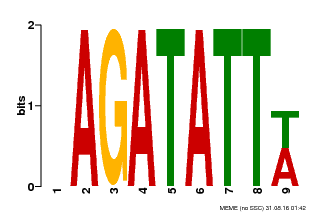
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019454353.1 | 1e-126 | PREDICTED: protein REVEILLE 6-like isoform X1 | ||||
| Refseq | XP_021643238.1 | 1e-126 | protein REVEILLE 6-like | ||||
| Swissprot | Q8H0W3 | 1e-105 | RVE6_ARATH; Protein REVEILLE 6 | ||||
| TrEMBL | A0A061G0F6 | 1e-125 | A0A061G0F6_THECC; Homeodomain-like superfamily protein isoform 1 | ||||
| TrEMBL | B9RIS7 | 1e-125 | B9RIS7_RICCO; DNA binding protein, putative | ||||
| STRING | EOY23325 | 1e-125 | (Theobroma cacao) | ||||
| STRING | XP_002513646.1 | 1e-125 | (Ricinus communis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52660.2 | 3e-91 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0277s0041.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



