 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0327s0019.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1294aa MW: 143713 Da PI: 8.9723 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.9 | 0.00016 | 1201 | 1223 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk+F ++ +L +H r+H
Kalax.0327s0019.1.p 1201 CPvtGCGKTFFSHKYLLQHRRVH 1223
9999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 10.9 | 0.0014 | 1259 | 1285 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y C Cg++F+ s++ rH r+ H
Kalax.0327s0019.1.p 1259 YMCAepGCGQTFRFVSDFSRHKRKtgH 1285
789999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.7E-17 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.478 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 8.8E-15 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 1.3E-46 | 193 | 362 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.078 | 196 | 362 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 8.79E-26 | 211 | 378 | No hit | No description |
| Pfam | PF02373 | 2.9E-37 | 226 | 345 | IPR003347 | JmjC domain |
| SMART | SM00355 | 26 | 1176 | 1198 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.05 | 1199 | 1228 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.23 | 1199 | 1223 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 8.09E-6 | 1201 | 1235 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.9E-5 | 1201 | 1222 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1201 | 1223 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.7E-8 | 1223 | 1251 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0014 | 1229 | 1253 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.471 | 1229 | 1258 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1231 | 1253 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.05E-9 | 1239 | 1284 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.3E-9 | 1252 | 1282 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.845 | 1259 | 1290 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 1.1 | 1259 | 1285 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1261 | 1285 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1294 aa Download sequence Send to blast |
MSGKQQQAEV FQWLRALPVA PEYRPTLDEF QDPIAYIFKI EKEASRYGIC KIVPPLPPAP 60 RKTVVANFNK SVPNGSGAKP TFTTRQQQIG FCPRRGRPVQ RPVWQSGEFY TFQEFEAKAR 120 AFERSYLKKC GKKAAVAAAA GAGLSALEIE TLYWKASADK PFSVEYANDM PGSAFAAASG 180 TGRRPREGGE GMTVGDTAWN MKGVARSTMS LLRFMKEDIA GVTSPMVYVA MLFSWFAWHV 240 EDHDLHSLNY LHTGAGKTWY GVPKEAAAGF EEVVRVHGYG GEINPLVTFA TLGEKTTVMS 300 PEVFVKAGIP CCRLVQNPGE FVVTFPRAYH SGFSHGFNCG EASNIATPYW LAVAKEAAVR 360 RASINYPPMV SHFQLLYDLA FALCSSLPMG VHSQPRSSRL KQKRKGVGET VVKQLFVEDI 420 KESNRLLQLL GEGLPVVLRP LSSAVFSIYS KSRIQCLLKS RSSSHLSLCY QKETVTSIRN 480 NLAASDINSG TNGRLPTVIG LPAVGSKFCS TFDGNSRSLP NESTEPQLST SSRWVTDYEN 540 SNAIQDDGAS DKKLFSCVTC GILCFSCVAI IQPAEVAARY LMASDCSFFS DSENAAKKTS 600 EELADSGRGV NTLDHTYSSG WMGRRAHNGK SNHIEEEYQS DEANSDREEQ PRTTALDLLA 660 SAYGNSSDSE NDTRLDSDPV YADGPNFSKI SSDSDFECND SDIRDPLQEN DFRWESSPRL 720 HSEGETTRFH IADSDGENHY RRAFQNRNST YDSPSFNLDK LASMGSNLAS LQSDKDSLRM 780 HVFCLEHAIE VKKKLKQIGG VNMLLLCHPD YPRVEAEAKS VQEELRLDHC WNDTAFRESS 840 EDDERCMAAA LESAEDVSSN GDWTVKLGIN LFYSASLSRS PLYSKQMPYN SIVYKAFGRV 900 PPLEAPAKSD GHTKGNSRKK KIVLAGKWCG KPWMSNQVHP LLLPPPPALE ETQQEHLHAH 960 ASASAKTDAA DSDSIQPVKS SKARRDDSSA TTSTPLAGSK RQIVVYSRSN KKTKRAETGL 1020 SVSASDGGDA DAGENLSNQQ AKLHVRSKHK IEGNKKPLEE ETRILYYSSK HVAKGDLKST 1080 KETTSLRDLY KPNFKPFGAD LDDKREGGLS TRLRRRNTKP ELEHEGKVLV KKERKPNKTP 1140 ANSKEAKIEK VDAKPRKSKV SEAARSRKKK DEEGEFQCDK EGCTMSFSLK QELALHKRDI 1200 CPVTGCGKTF FSHKYLLQHR RVHEDARPLK CPWKGCKMTF KWAWARTEHI RVHTGVRPYM 1260 CAEPGCGQTF RFVSDFSRHK RKTGHSVKRN IKC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 3e-76 | 7 | 386 | 2 | 356 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 3e-76 | 7 | 386 | 2 | 356 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
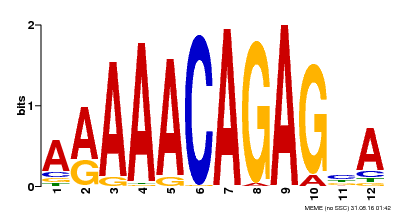
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010663055.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | F6GTM2 | 0.0 | F6GTM2_VITVI; Uncharacterized protein | ||||
| STRING | VIT_17s0000g09980.t01 | 0.0 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0327s0019.1.p |



