 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0336s0030.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 253aa MW: 28907 Da PI: 8.8623 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 83.8 | 1e-26 | 9 | 61 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TT..SEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsst..gklyeyss 51
k+ienk nrqvtfskRr+g++KKA ELSvLCda++a+i+fs + +klye++s
Kalax.0336s0030.1.p 9 KKIENKVNRQVTFSKRRTGLFKKATELSVLCDAQIAIIVFSGDrpDKLYEFAS 61
68***************************************874579***986 PP
| |||||||
| 2 | K-box | 79.9 | 6.1e-27 | 86 | 173 | 12 | 99 |
K-box 12 akaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkkle 99
++a+s ++e++kL+++ e Lqr+qR+llGedLe+Ls+keL+ Le+q+e sl + R +K++++ e +e l+k+e+ l e+nk+L++k +
Kalax.0336s0030.1.p 86 REAQSAYHEVQKLQAKCEALQRTQRNLLGEDLEPLSIKELHALEKQIEVSLAQTRLRKTQVVNERMESLRKQERHLGEINKKLKMKKM 173
57899*******************************************************************************9976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 6.7E-34 | 1 | 62 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 27.656 | 1 | 63 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 3.41E-36 | 2 | 74 | No hit | No description |
| SuperFamily | SSF55455 | 5.49E-29 | 2 | 80 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 9.3E-21 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 3.1E-24 | 10 | 59 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 9.3E-21 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 9.3E-21 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 1.0E-23 | 86 | 171 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.569 | 88 | 178 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048437 | Biological Process | floral organ development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 253 aa Download sequence Send to blast |
MGRGRIEHKK IENKVNRQVT FSKRRTGLFK KATELSVLCD AQIAIIVFSG DRPDKLYEFA 60 SPGQRIGKIL ERYQSMVAPP PGGLHREAQS AYHEVQKLQA KCEALQRTQR NLLGEDLEPL 120 SIKELHALEK QIEVSLAQTR LRKTQVVNER MESLRKQERH LGEINKKLKM KKMEIEAEGE 180 TYHALWNTYN ASASAGTNFP LQPTHPNLME CEMAEPVLEM GYPQNHYVRE EGTTSRANNV 240 VDELNFIQGW HC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 6e-19 | 78 | 171 | 3 | 100 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 6e-19 | 78 | 171 | 3 | 100 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 6e-19 | 78 | 171 | 3 | 100 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 6e-19 | 78 | 171 | 3 | 100 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in flower development. {ECO:0000250|UniProtKB:Q0HA25}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00315 | DAP | Transfer from AT2G45650 | Download |
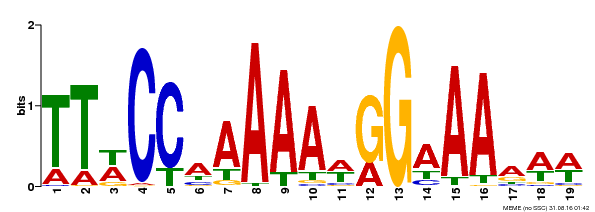
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001280892.1 | 3e-86 | agamous-like MADS-box protein AGL6 | ||||
| Swissprot | Q8LLR1 | 6e-85 | MADS3_VITVI; Agamous-like MADS-box protein MADS3 | ||||
| TrEMBL | O82699 | 8e-85 | O82699_MALDO; MADS domain class transcription factor | ||||
| STRING | XP_008342931.1 | 1e-85 | (Malus domestica) | ||||
| STRING | XP_008347113.1 | 1e-85 | (Malus domestica) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45650.1 | 2e-83 | AGAMOUS-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0336s0030.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



