 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0495s0013.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 498aa MW: 54479.9 Da PI: 4.9772 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 41.9 | 1.6e-13 | 170 | 193 | 3 | 26 |
S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 3 telCrffartGtCkyGdrCkFaHg 26
e C+f++rtG CkyG +CkF+H+
Kalax.0495s0013.1.p 170 AEDCNFYMRTGSCKYGLSCKFNHP 193
689********************9 PP
| |||||||
| 2 | zf-CCCH | 22.3 | 2.3e-07 | 217 | 240 | 4 | 27 |
---SGGGGTS--TTTTT-SS-SSS CS
zf-CCCH 4 elCrffartGtCkyGdrCkFaHgp 27
+C++++ G CkyG C+F H++
Kalax.0495s0013.1.p 217 TECKYYLSAGGCKYGKACRFEHSR 240
58******************9985 PP
| |||||||
| 3 | zf-CCCH | 34.2 | 4.3e-11 | 261 | 285 | 2 | 26 |
-S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 2 ktelCrffartGtCkyGdrCkFaHg 26
++++C++++r+G Cky ++C+F+H+
Kalax.0495s0013.1.p 261 GEKECPYYMRNGSCKYAASCRFNHP 285
7899********************9 PP
| |||||||
| 4 | zf-CCCH | 33.3 | 8.1e-11 | 384 | 410 | 1 | 27 |
--S---SGGGGTS--TTTTT-SS-SSS CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaHgp 27
+++++C ff+rtG Cky ++CkF+H++
Kalax.0495s0013.1.p 384 PGQPECTFFLRTGDCKYRSTCKFHHPK 410
5789*********************96 PP
| |||||||
| 5 | zf-CCCH | 24.8 | 3.8e-08 | 430 | 455 | 2 | 27 |
-S---SGGGGTS--TTTTT-SS-SSS CS
zf-CCCH 2 ktelCrffartGtCkyGdrCkFaHgp 27
++++C +++ +G+Ck+G++C++ H+p
Kalax.0495s0013.1.p 430 GQSICTHYRHYGICKFGPKCRYDHPP 455
7899********************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00356 | 2.1E-7 | 167 | 194 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 14.596 | 167 | 195 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 4.32E-6 | 168 | 196 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 7.9E-11 | 170 | 193 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 2.5E-5 | 173 | 192 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 0.0012 | 213 | 240 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 13.892 | 213 | 241 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 1.2E-5 | 217 | 239 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 1.57E-5 | 217 | 242 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 2.9E-5 | 219 | 241 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 1.0E-5 | 259 | 286 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 14.896 | 259 | 287 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 2.88E-5 | 261 | 286 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 5.8E-8 | 261 | 285 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 2.8E-4 | 265 | 284 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 3.8E-4 | 383 | 410 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 14.051 | 383 | 411 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 1.1E-6 | 385 | 410 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 3.92E-5 | 387 | 411 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 6.1E-4 | 428 | 455 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 15.112 | 428 | 456 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 1.0E-5 | 430 | 455 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 3.53E-5 | 431 | 455 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 498 aa Download sequence Send to blast |
METPEPIPSP DSDQHQPPIH DSQPPLPDPS DDDPKSPPPA PPTEELQAMI LKEDEAPGDG 60 DGDGGRVNGV SERKDEADGG FGGVGGGGSE NEGVAESETE RHGEKEKVEE VVKDGWSDDQ 120 QGRVDGGVEE TGDGYPVDGW SIGNGEGEPD WKEEAQGRDM QVQYPLRPGA EDCNFYMRTG 180 SCKYGLSCKF NHPLRRTNQT VKPRVKERED SSERPMTECK YYLSAGGCKY GKACRFEHSR 240 ETPSVVPVLE YNFLGLPIRP GEKECPYYMR NGSCKYAASC RFNHPDPTAG SDTPSGYAND 300 EHASSWSPSR PSNKVASFVP GSYTSAQGIH PQISEWNGYQ GPVFTAEKSF RPPPAFGLNI 360 PPMAGSAYSH FPSQLASEEF PERPGQPECT FFLRTGDCKY RSTCKFHHPK DRNVLPLCAM 420 TDKGLPLRPG QSICTHYRHY GICKFGPKCR YDHPPNIQQS EPSAMTGLGQ PHHPFSNFAS 480 PEDASRLSSM GEGTITL* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00582 | DAP | Transfer from AT5G63260 | Download |
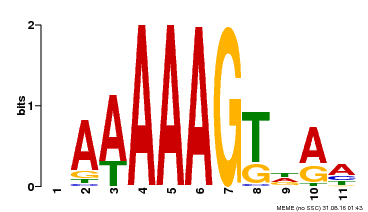
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028781830.1 | 1e-134 | zinc finger CCCH domain-containing protein 67-like isoform X3 | ||||
| Swissprot | Q5RJC5 | 1e-111 | C3H67_ARATH; Zinc finger CCCH domain-containing protein 67 | ||||
| STRING | EOY22360 | 1e-130 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G63260.1 | 1e-113 | C3H family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0495s0013.1.p |



