 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0514s0014.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 360aa MW: 40666.1 Da PI: 7.0763 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 171.2 | 3.2e-53 | 10 | 140 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvka.eekewyfFskrdkkyatgkrknratksgyWkatg 87
+ppGfrFhPtdeel+++yL+kkv+g++++l +vi+e+d++k+ePwdL+ k ++ + ++ewyfFs++dkky+tg+r+nrat++g+Wkatg
Kalax.0514s0014.1.p 10 VPPGFRFHPTDEELLNYYLRKKVSGEEIDL-DVIREIDVNKLEPWDLKdKcRIGSgPQNEWYFFSHKDKKYPTGTRTNRATTAGFWKATG 98
69****************************.9***************943444443456******************************* PP
NAM 88 kdkevlsk.kgelvglkktLvfykgrapkgektdWvmheyrl 128
+dk+v+++ +++ gl+ktLvfy+grap+g+ktdW+mheyrl
Kalax.0514s0014.1.p 99 RDKAVHHNgSSRRLGLRKTLVFYTGRAPHGQKTDWIMHEYRL 140
*******866778***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 2.75E-59 | 5 | 168 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.045 | 10 | 168 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.2E-28 | 11 | 140 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003002 | Biological Process | regionalization | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009834 | Biological Process | plant-type secondary cell wall biogenesis | ||||
| GO:0010455 | Biological Process | positive regulation of cell fate commitment | ||||
| GO:0048829 | Biological Process | root cap development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 360 aa Download sequence Send to blast |
MSGSSGQLSV PPGFRFHPTD EELLNYYLRK KVSGEEIDLD VIREIDVNKL EPWDLKDKCR 60 IGSGPQNEWY FFSHKDKKYP TGTRTNRATT AGFWKATGRD KAVHHNGSSR RLGLRKTLVF 120 YTGRAPHGQK TDWIMHEYRL DDHHNITHNT TLDHQEDDGW VVCRVFKKKN NPRSDYEQDL 180 KNPNTSKNDE DESLNQRAWP PSSYNNTNNG SSTPMILMQP PHMNYPSAII NTSMHLPQLF 240 SPEANVPTFM SPIITSNDNN NADVECSHSL LRLMSSSSSP AAASGLLSLN NNTGNSSDWS 300 FLDKLLATNN ISQHIQLLDH HHHYNAPPSF QLVDYVGASA HKTHLDCFGC ESDSLKYSK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| 3swm_B | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| 3swm_C | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| 3swm_D | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| 3swp_A | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| 3swp_B | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| 3swp_C | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| 3swp_D | 8e-54 | 7 | 171 | 17 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription regulator. Together with BRN1 and BRN2, regulates cellular maturation of root cap. Represses stem cell-like divisions in the root cap daughter cells, and thus promotes daughter cell fate. Inhibits expression of its positive regulator FEZ in a feedback loop for controlled switches in cell division plane. Promotes the expression of genes involved in secondary cell walls (SCW) biosynthesis. {ECO:0000269|PubMed:19081078, ECO:0000269|PubMed:20197506}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00250 | DAP | Transfer from AT1G79580 | Download |
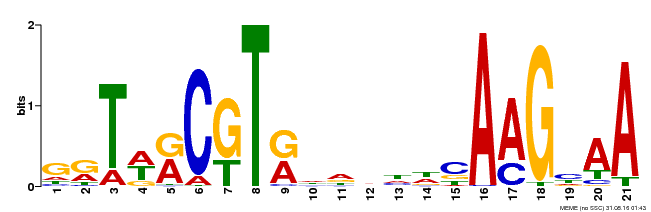
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By FEZ in oriented-divised root cap stem cells. {ECO:0000269|PubMed:19081078}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002316163.2 | 1e-129 | protein SOMBRERO | ||||
| Refseq | XP_011014561.1 | 1e-129 | PREDICTED: protein SOMBRERO-like | ||||
| Refseq | XP_011014569.1 | 1e-129 | PREDICTED: protein SOMBRERO-like | ||||
| Refseq | XP_024466050.1 | 1e-129 | protein SOMBRERO | ||||
| Swissprot | Q9MA17 | 1e-106 | SMB_ARATH; Protein SOMBRERO | ||||
| TrEMBL | A0A3N7HAQ9 | 1e-127 | A0A3N7HAQ9_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0010s18420.1 | 1e-128 | (Populus trichocarpa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G79580.3 | 2e-88 | NAC family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0514s0014.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



