 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0534s0004.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 652aa MW: 73604.1 Da PI: 6.5789 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 132.3 | 5.5e-41 | 52 | 153 | 6 | 105 |
DUF822 6 kptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl..eeaeaagssasaspess 93
k++++E+E++k+RER+RRa++++++aGLR++Gn++lp+raD+n+Vl+AL+r+AGw+ve+DGttyr++ p+ ++ea + sp s
Kalax.0534s0004.2.p 52 KKREREKERTKLRERHRRAVTSRMLAGLRQYGNFPLPARADMNDVLAALARQAGWTVEADGTTYRHSALPNpvVQMEAYPVWSIDSPVSA 141
6789***************************************************************99996666666666666666666 PP
DUF822 94 lqsslkssalas 105
+++ s++l
Kalax.0534s0004.2.p 142 TSLGKASCSLDG 153
665555555544 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.2E-40 | 52 | 153 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 3.66E-146 | 215 | 648 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 4.2E-158 | 218 | 649 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.5E-76 | 242 | 612 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 255 | 269 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 276 | 294 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 298 | 319 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 391 | 413 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 464 | 483 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 498 | 514 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 515 | 526 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 533 | 556 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.6E-49 | 571 | 593 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 652 aa Download sequence Send to blast |
MKTGNEIGAI QHLDPTHHHH QQQQPPSRRP RGFAASAAIH APGSTSACVK GKKREREKER 60 TKLRERHRRA VTSRMLAGLR QYGNFPLPAR ADMNDVLAAL ARQAGWTVEA DGTTYRHSAL 120 PNPVVQMEAY PVWSIDSPVS ATSLGKASCS LDGLHSLQVT LRGHEGTNLS PVASFDSAVI 180 SERDTGRSID TYTSNSLEAD QLLHSTRFGE DQSDFAGTPY IPVYIALAPD IINNLGQLID 240 PDGIRQELSH IKSLNADGVV VLCWWGIVEN WIPQKYEWMG YKLLFNIIRE FKLKLQVVMA 300 FHDYGIDDSG NVLIPLPQWV LDIGRENPDI FFTDRKGERN MECLTLGIDK ERALNKRTGI 360 EVYYDFMRNF RTEFDDLFSE GIVYAVEIGL GASGELKYPA FSERSGWRYP GIGEFQCYDK 420 YLQQSLKLAA KLRGHPFWAK APDNCGHYNS KPFETGFFHE RGDYDSYYGR FFLQWYTQCL 480 IDHADNILSL ASLIFEGTHL VIKIPAIFWW YKTVSHAAEL AAGYYNPTNK DGYSPLFEVL 540 NKHGVTVKFA HSGLLLTSQE SDEALADPES LSWQVLNSAW ERGLTVAGEN PIGLYDKEGY 600 MRVVETAKPR NDPDRHRLSF FVHQYASPLF QTAICVSELD YFIKCMHGEK L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wds_A | 1e-119 | 220 | 621 | 11 | 414 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
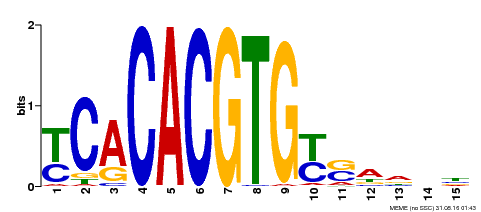
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002270680.1 | 0.0 | PREDICTED: beta-amylase 8 isoform X1 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A067JVR7 | 0.0 | A0A067JVR7_JATCU; Beta-amylase | ||||
| STRING | EOY25398 | 0.0 | (Theobroma cacao) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0534s0004.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



